Wolfram Data Repository
Immediate Computable Access to Curated Contributed Data
Dataset of the transcriptional regulation network of Escherichia coli
Originator:
Shai Shen-Orr, under the supervision of Dr. Uri Alon
Weizmann Institute of Science
The expression of genes must be carefully controlled to occur under the correct developmental and environmental conditions. To do so, expressed proteins regulate the transcription of genes, forming links of activation and inhibition constituting networks of coordination. This data set contains the regulated operons of E. coli, the transcription factors regulating those operons, and whether that regulation activates or inhibits expression. (Operons are one or more genes transcribed on the same DNA). With this regulatory graph, we can find common graph motifs that suggest particular regulatory relationships and allows us to understand aspects of regulatory dynamics more clearly.
Retrieve the resource:
| In[1]:= |
| Out[1]= |  |
Retrieve the default content, a Dataset of the regulatory network:
| In[2]:= |
| Out[2]= | 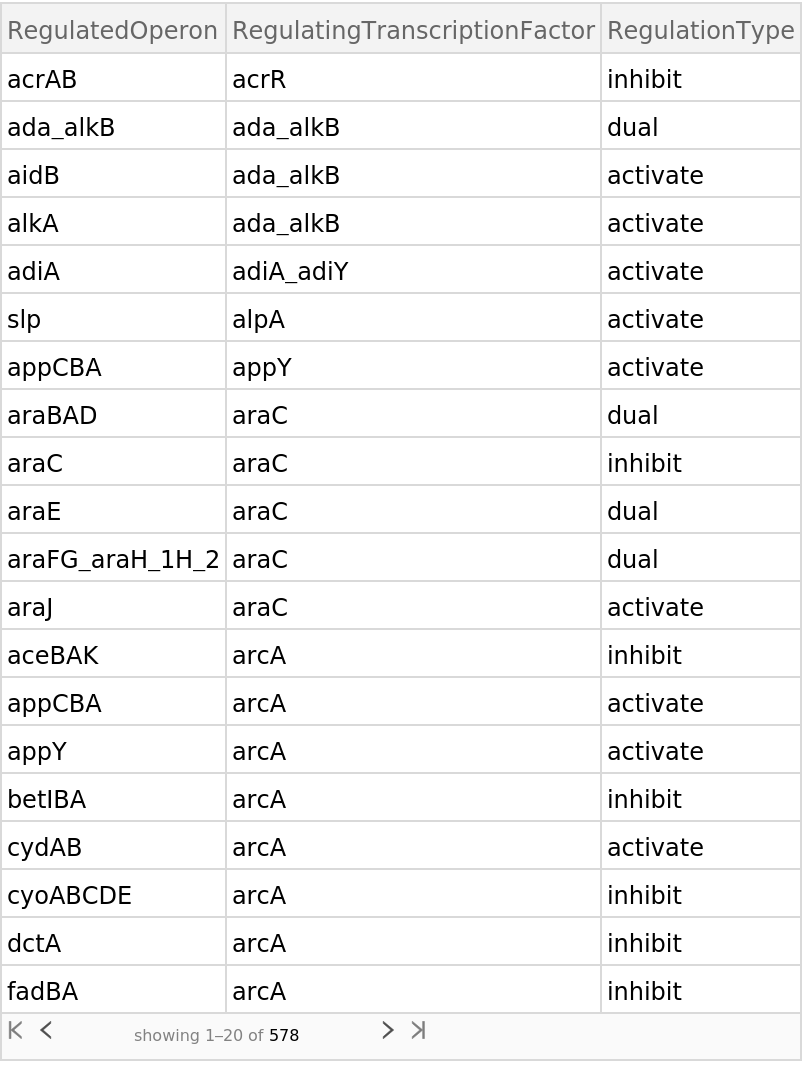 |
Visualize a graph of this regulatory network:
| In[3]:= | ![Graph[Normal[
DirectedEdge[#["RegulatingTranscriptionFactor"], #[
"RegulatedOperon"]] & /@ ResourceData[
"Transcriptional Regulation Network of Escherichia coli"]],
EdgeStyle -> Normal[(#["RegulatingTranscriptionFactor"] -> #[
"RegulatedOperon"]) -> #["RegulationType"] /. {"activate" ->
Red, "inhibit" -> Blue, "dual" -> Purple} & /@ ResourceData[
"Transcriptional Regulation Network of Escherichia coli"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/35d/35db07c8-c154-4674-85b9-e2da8b07606e/6450cc87045a4a79.png) |
| Out[3]= | 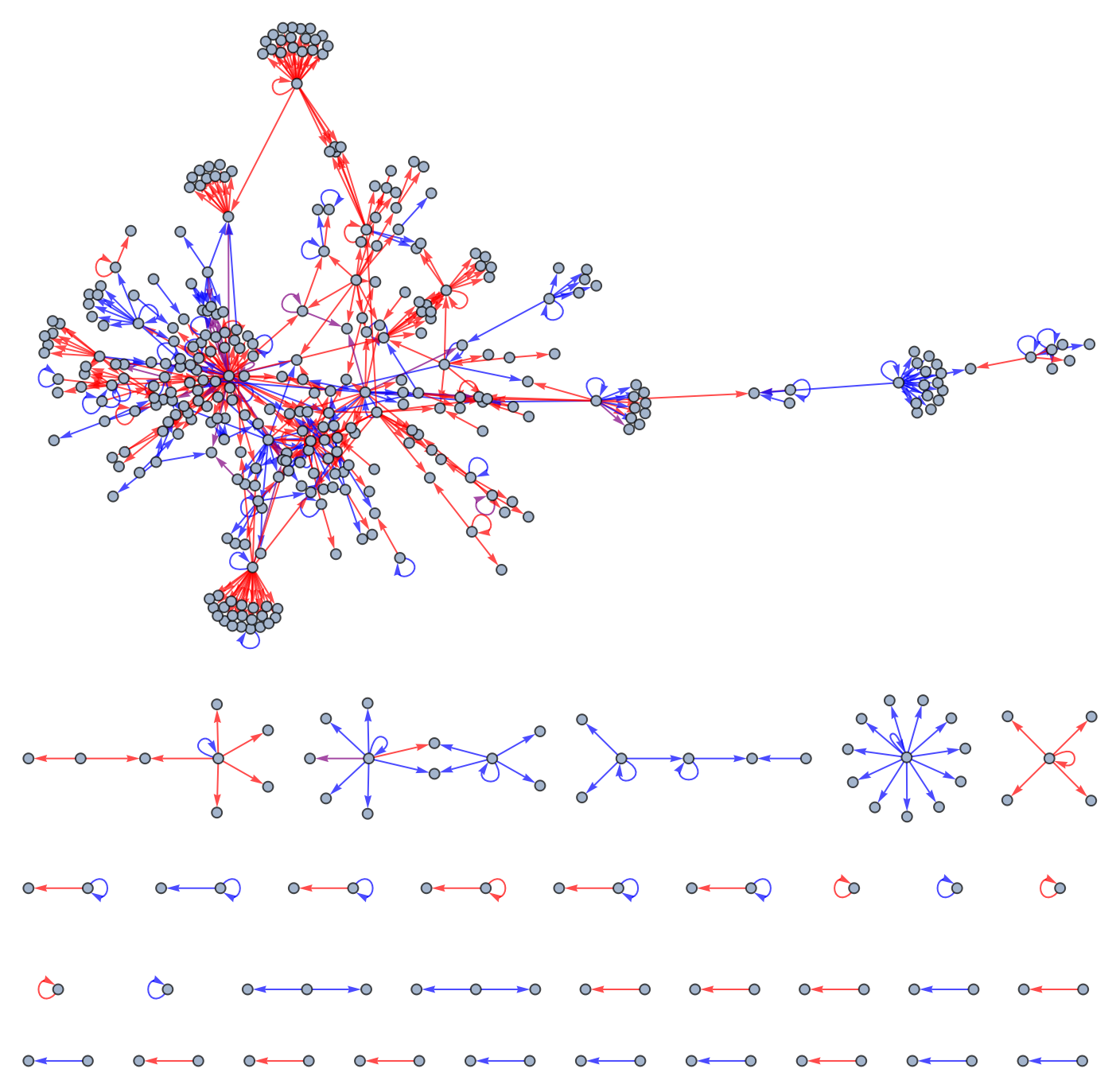 |
Wolfram Research, "Transcriptional Regulation Network of Escherichia coli" from the Wolfram Data Repository (2018)