Wolfram Data Repository
Immediate Computable Access to Curated Contributed Data
Categorization of the pathogenicity of 89% of 71 million possible human missense variants
| "Chromosome" | string including "Chromosome" and a number from 1 to 22 or a letter M,X,or Y. |
| "RawDataSize" | total rows for each chromosome in the original database |
| "TotalPositions" | total positions for each chromosome |
| "TotalMutations" | total missense mutations for each chromosome |
| "Genome" | the genome build |
| "TotalUniprotID" | total uniprot IDs |
| "TotalTranscriptID" | total transcript IDs |
| "TotalAminoAcidVariations" | total amino acid changes |
| "PathogenicityQuartiles" | pathogenicity Quartiles 1/4, 2/4, 3/4 |
| "PathogenicityMean" | pathogenicity Mean |
| "PathogenicityLikelyBenign" | percentage of "likely_benign" classifications |
| "PathogenicityLikelyPathogenic" | percentage of "likely_pathogenic" classifications |
| "PathogenicityAmbiguous" | percentage of "ambiguous" classifications |
| "Chromosome" | string including "Chromosome" and a number from 1 to 22 or a letter M,X,or Y. |
| "Position" | genome position (1-based) |
| "ReferenceNucleotide" | reference nucleotide (GRCh38.p13 for hg38) |
| "AlternativeNucleotide" | alternative nucleotide |
| "Genome" | genome build |
| "UniprotID" | UniProtKB accession number of the protein in which the variant induces a single amino-acid substitution (UniProt release 2021_02) |
| "TranscriptID" | Ensembl transcript ID from GENCODE V32 (hg38) |
| "AminoAcidVariation" | Amino acid change induced by the alternative allele,in the format: Reference aminoacid-POS_aa-Alternative amino acid |
| "Pathogenicity" | predicted probability of a variant being clinically pathogenic |
| "Classification" | derived using the following thresholds: "likely_benign" for Pathogenicity < 0.34; "likely_pathogenic" for Pathogenicity > 0.564; and "ambiguous" otherwise |
| In[1]:= |
| Out[1]= | 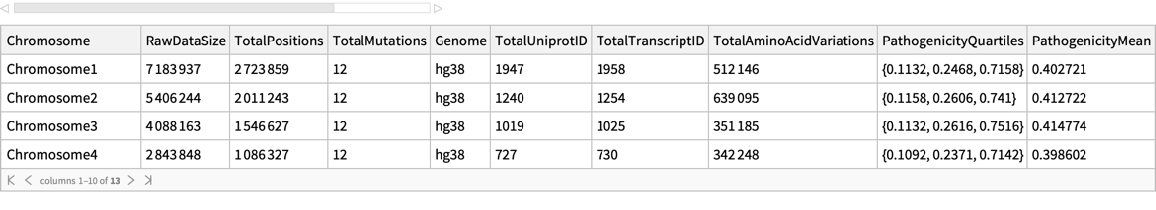 |
Retrieve the data for ChromosomeX:
| In[2]:= |
| Out[21]= | 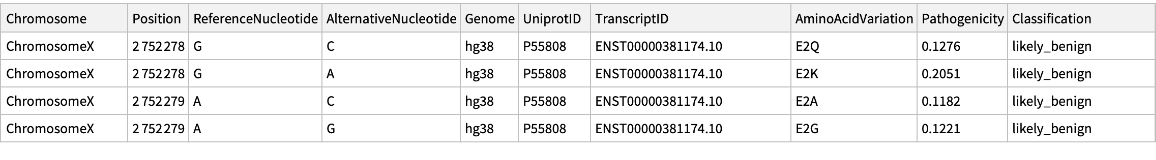 |
Get a random sample of the Positions in ChromosomeX:
| In[22]:= |
| Out[22]= |
Get a random sample of the transcriptIDs in ChromosomeX:
| In[23]:= |
| Out[23]= |
Get a distribution of the ChromosomeX pathogenicity level for all possible missense variations:
| In[24]:= | ![BoxWhiskerChart[chrX[All, #Pathogenicity &], "Mean", PlotLabel -> "Pathogenicy level distribution", FrameLabel -> {"ChromosomeX", "Pathogenicity level"}]](https://www.wolframcloud.com/obj/resourcesystem/images/225/225dffa1-1f90-49d1-912c-d751dd984043/32544d2671167aa4.png) |
| Out[24]= | 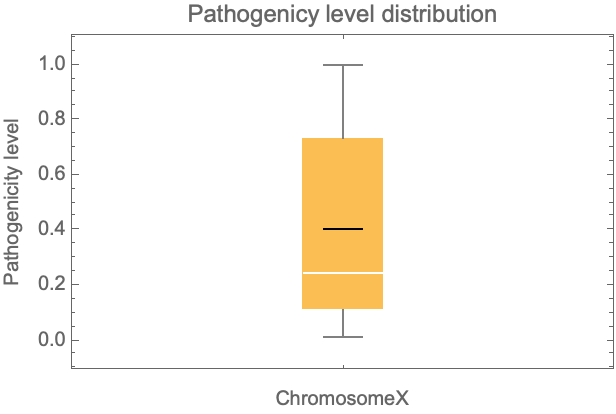 |
Get a summary of the classification of the ChromosomeX pathogenicity level for all possible missense variations:
| In[25]:= |
| Out[25]= | 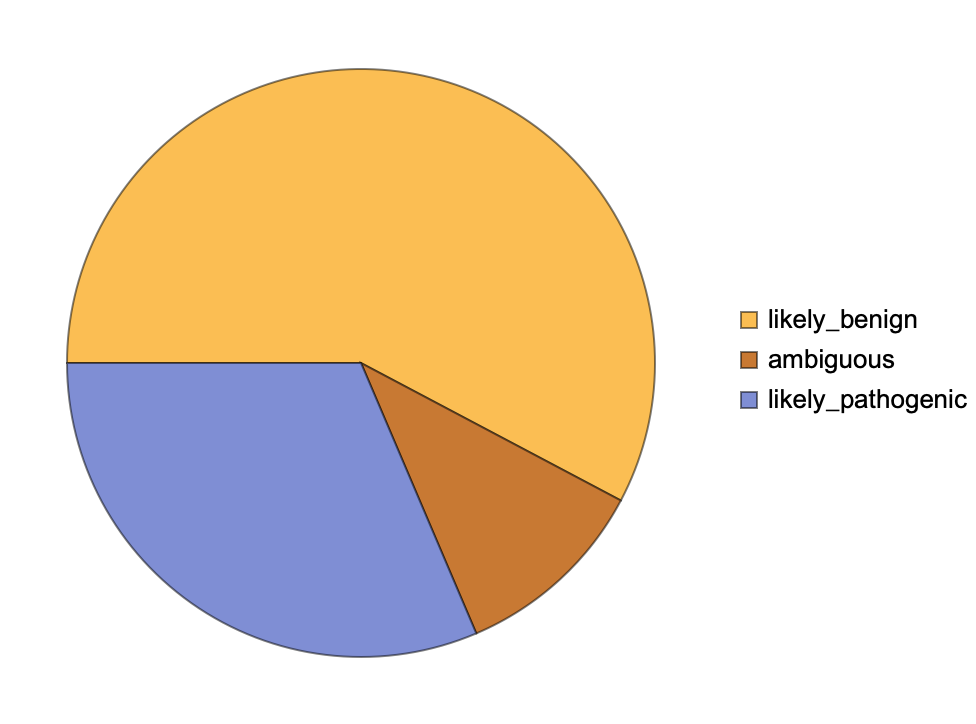 |
Get the pathogenicity information associated to ChromosomeX in position 71765227:
| In[26]:= | ![PathogenicityData[chromosome_String, mutationPosition_Integer, "Pathogenicity"] := Block[{rawData0},
rawData0 = ResourceData[\!\(\*
TagBox["\"\<Alpha Missense\>\"",
#& ,
BoxID -> "ResourceTag-Alpha Missense-Input",
AutoDelete->True]\), chromosome][
Select[MatchQ[#[[2]], mutationPosition] &]];
Dataset[
Association[{"Position" -> #[[2]], "Mutation" -> {#[[3]] -> #[[4]]}, "TranscriptID" -> #[[7]], "AminoAcidVariation" -> #[[8]], "Pathogenicity" -> #[[9]], "Classification" -> #[[10]]}] & /@ Normal[rawData0]]
]](https://www.wolframcloud.com/obj/resourcesystem/images/225/225dffa1-1f90-49d1-912c-d751dd984043/48ab4cea46a83d84.png) |
| In[27]:= |
| Out[27]= | 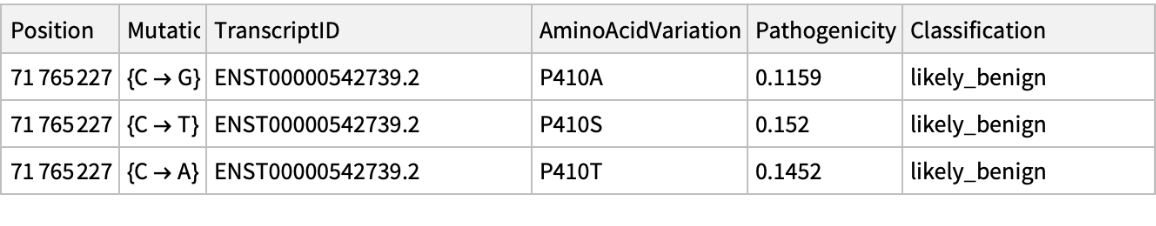 |
Get the pathogenicity information associated to ChromosomeX for a list of positions:
| In[28]:= | ![PathogenicityData[chromosome_String, mutationPosition_List, "Pathogenicity"] := Block[{rawData0},
rawData0 = ResourceData[\!\(\*
TagBox["\"\<Alpha Missense\>\"",
#& ,
BoxID -> "ResourceTag-Alpha Missense-Input",
AutoDelete->True]\), chromosome][
Select[MemberQ[mutationPosition, #[[2]]] &]];
Dataset[
Association[{"Position" -> #[[2]], "Mutation" -> {#[[3]] -> #[[4]]}, "TranscriptID" -> #[[7]], "AminoAcidVariation" -> #[[8]], "Pathogenicity" -> #[[9]], "Classification" -> #[[10]]}] & /@ Normal[rawData0]]
]](https://www.wolframcloud.com/obj/resourcesystem/images/225/225dffa1-1f90-49d1-912c-d751dd984043/4e1d489a9d6d4470.png) |
| In[29]:= |
| Out[29]= | 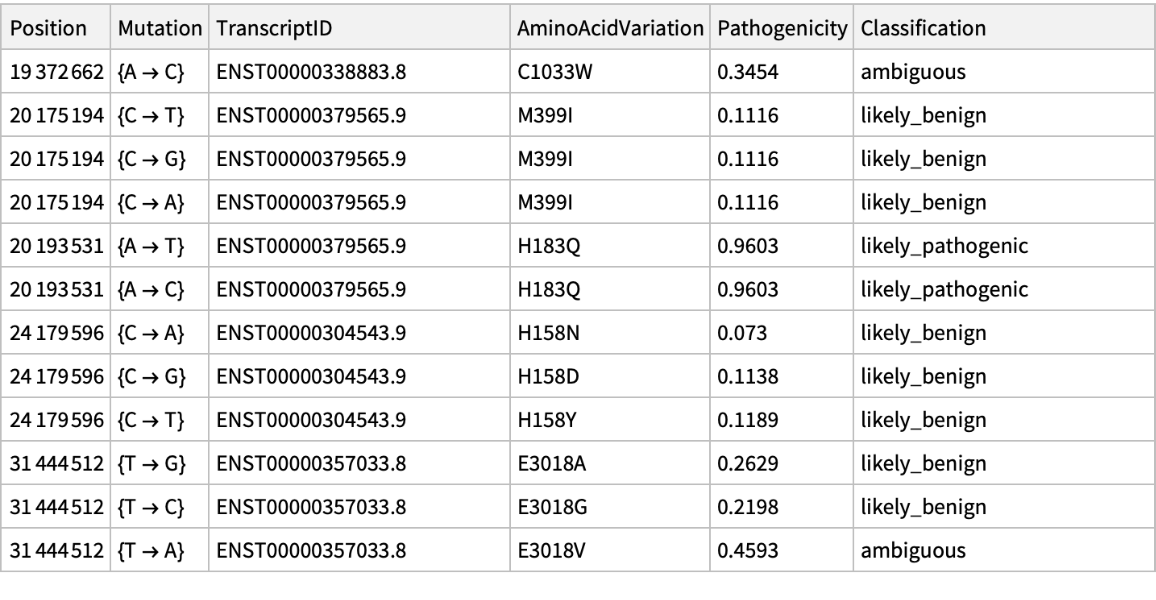 |
Get the pathogenicity distribution associated to all the positions of all the chromosomes:
| In[30]:= | ![data = Table[Normal[ResourceData[\!\(\*
TagBox["\"\<Alpha Missense\>\"",
#& ,
BoxID -> "ResourceTag-Alpha Missense-Input",
AutoDelete->True]\), "Chromosome" <> ToString[i]][
All, #Pathogenicity &]], {i, {Sequence @@ Range[1, 22], "M", "X",
"Y"}}];](https://www.wolframcloud.com/obj/resourcesystem/images/225/225dffa1-1f90-49d1-912c-d751dd984043/2bc88446bcbd0f12.png) |
| In[31]:= | ![BoxWhiskerChart[data, "Mean", ChartLabels -> {Sequence @@ Table["chr" <> ToString[i], {i, 1, 22, 1}], "chrM", "chrX", "chrY"}, PlotLabel -> "Pathogenicy level distribution", FrameLabel -> {"", "Pathogenicity level"}]](https://www.wolframcloud.com/obj/resourcesystem/images/225/225dffa1-1f90-49d1-912c-d751dd984043/33f3d2996fc5cab2.png) |
| Out[31]= | 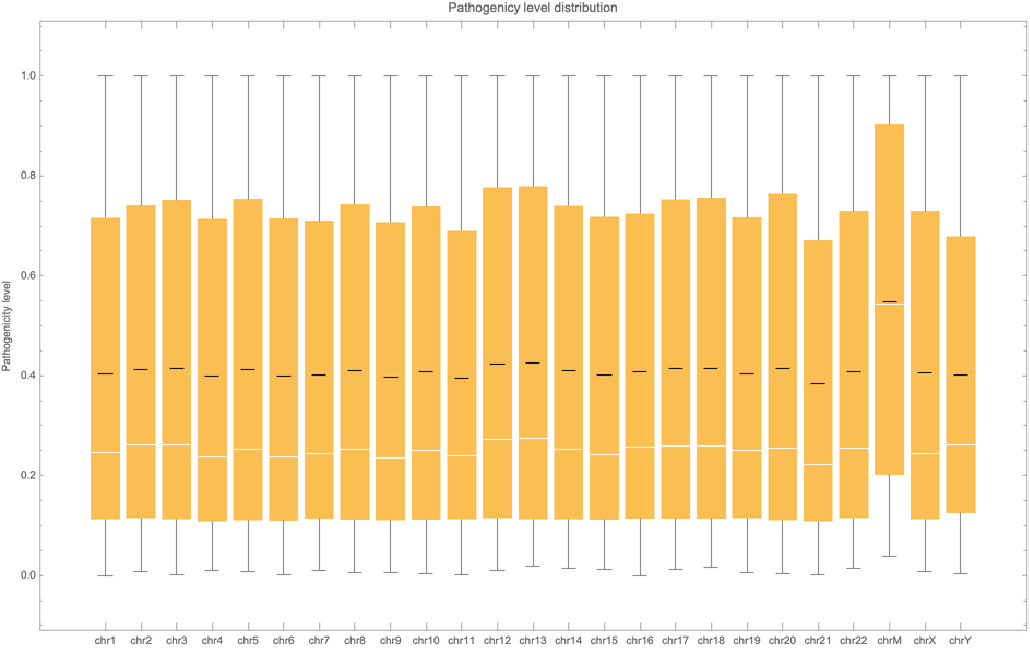 |
Wolfram Research, "Alpha Missense" from the Wolfram Data Repository (2024)