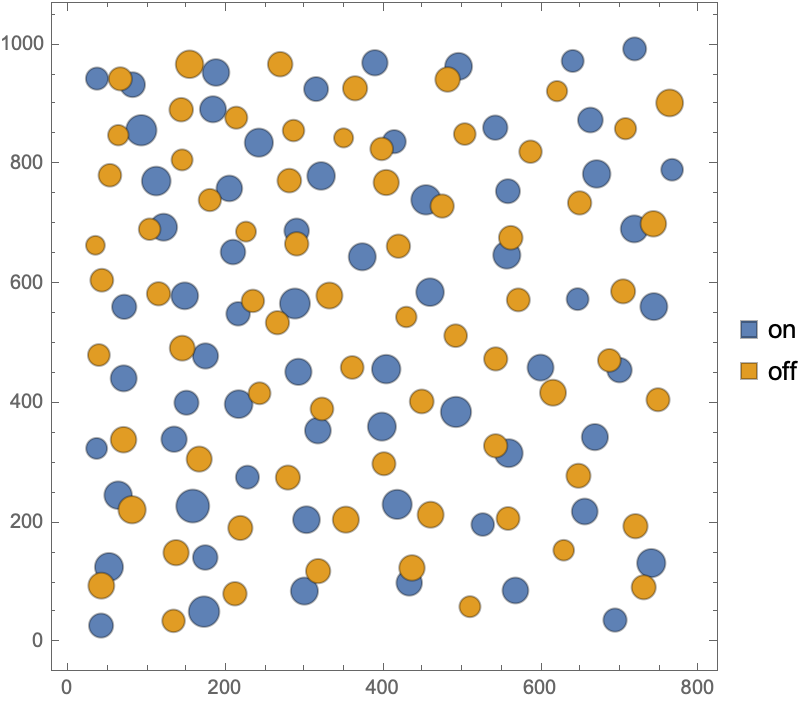
Locations of retinal ganglia cells annotated with on/off and area (in square microns) marks
Examples
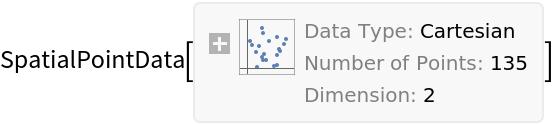
Basic Examples (1)
Summary of the spatial point data:
Visualizations (4)
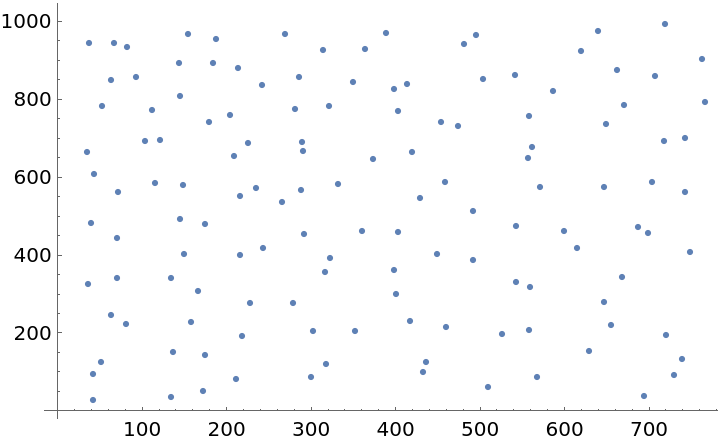
Plot the spatial point data:
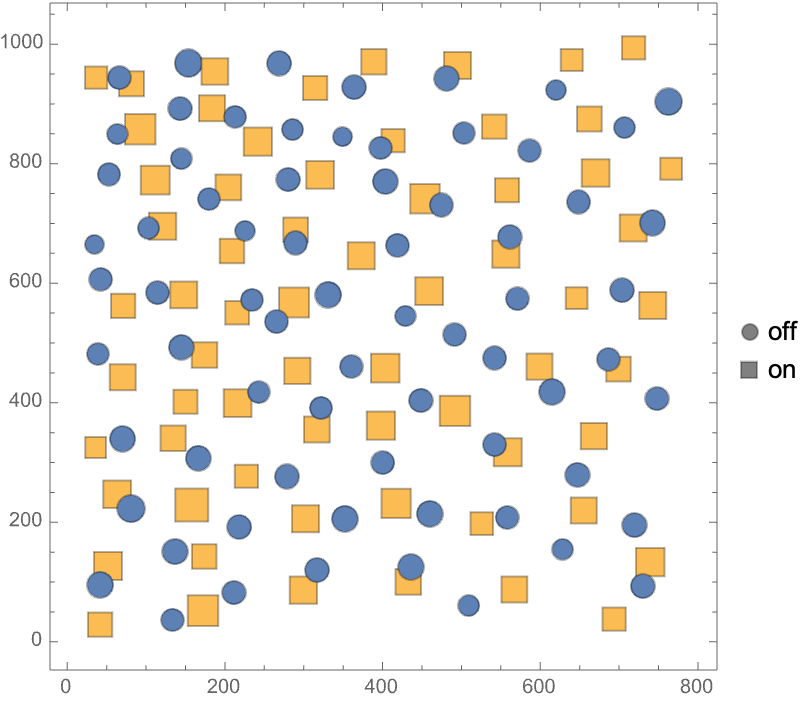
Visualize data with categorical annotations:
Visualize data with both annotations:
Visualize smooth point density:
Analysis (6)
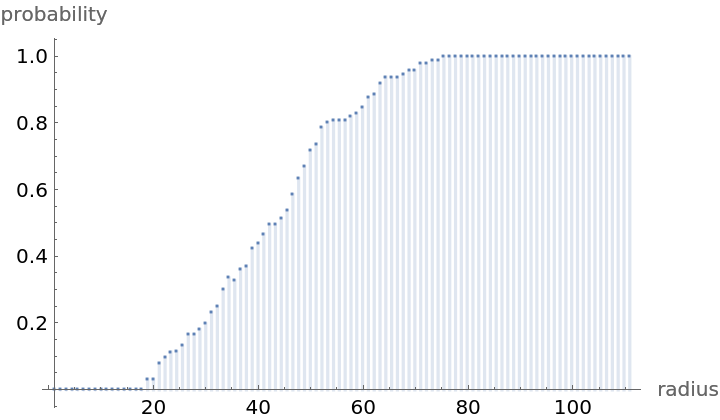
Compute probability of finding a point within given radius of an existing point - NearestNeighborG is the CDF of the nearest neighbor distribution:
NearestNeighborG as the CDF of nearest neighbor distribution can be used to compute the mean distance between a typical point and its nearest neighbor - the mean of a positive support distribution can be approximated via a Riemann sum of 1- CDF. To use Riemann approximation create the partition of the support interval from 0 to maxR into 100 parts and compute the value of the NearestNeighborG at the middle of each subinterval:
Now compute the Riemann sum to find the mean distance between a typical point and its nearest neighbor:
Account for scale and units:
Test for complete spatial randomness:
Fit a Poisson point process to data:
Bibliographic Citation
Gosia Konwerska,
"Sample Data: Beta Cells"
from the Wolfram Data Repository
(2022)
Data Resource History
Publisher Information
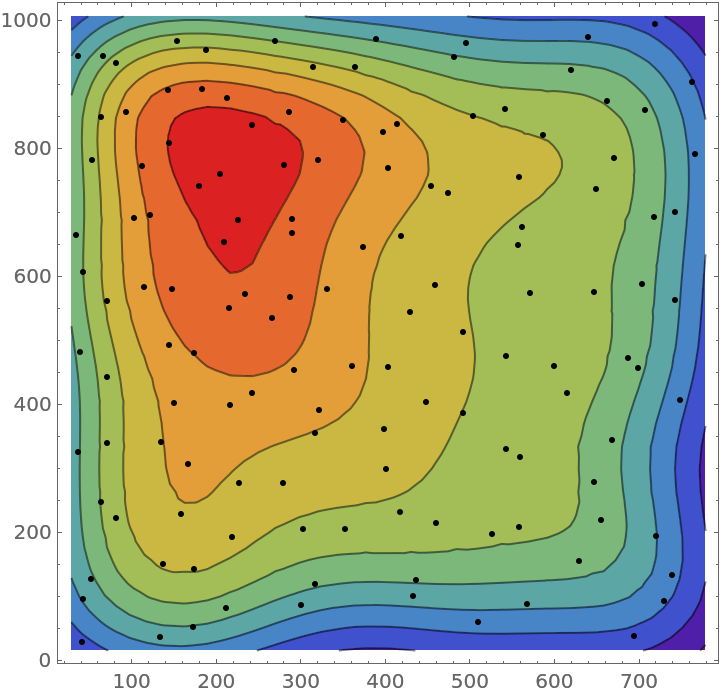
![Show[ContourPlot[density[{x, y}], {x, y} \[Element] ResourceData[\!\(\*
TagBox["\"\<Sample Data: Beta Cells\>\"",
#& ,
BoxID -> "ResourceTag-Sample Data: Beta Cells-Input",
AutoDelete->True]\), "ObservationRegion"], ColorFunction -> "Rainbow"], ListPlot[ResourceData[\!\(\*
TagBox["\"\<Sample Data: Beta Cells\>\"",
#& ,
BoxID -> "ResourceTag-Sample Data: Beta Cells-Input",
AutoDelete->True]\), "Locations"], PlotStyle -> Black]]](https://www.wolframcloud.com/obj/resourcesystem/images/34f/34f1c876-41b3-4ec7-adb7-efa9afa4262a/0cb3d1a93cb9adf5.png)

![step = maxR/100;
middles = Subdivide[step/2, maxR - step/2, 99];
values = nnG[middles];](https://www.wolframcloud.com/obj/resourcesystem/images/34f/34f1c876-41b3-4ec7-adb7-efa9afa4262a/25a9527b9c5861eb.png)

![Clear[\[Mu]];
EstimatedPointProcess[ResourceData[\!\(\*
TagBox["\"\<Sample Data: Beta Cells\>\"",
#& ,
BoxID -> "ResourceTag-Sample Data: Beta Cells-Input",
AutoDelete->True]\), "Data"], PoissonPointProcess[\[Mu], 2]]](https://www.wolframcloud.com/obj/resourcesystem/images/34f/34f1c876-41b3-4ec7-adb7-efa9afa4262a/522357db7641067a.png)