Details
The data is based on The Human Protein Atlas version 23.0 and Ensembl version 109.
Each dataset in the UMAP coordinates for cells is an
Association of the form
<|cluster id1 → {{cell id11, {UMAP_x11, UMAP_y11}}, {cell id12, {UMAP_x12, UMAP_y12}}, …, {cell id1N, {UMAP_x1N, UMAP_y1N}}}, …, cluster idM → {{cell idM1, {UMAP_xM1, UMAP_yM1}}, {cell idM2, {UMAP_xM2, UMAP_yM2}}, …, {cell idMK, {UMAP_xMK, UMAP_yMK}}}|>, where M, N, K are positive integers.
Uniform Manifold Approximation and Projection (UMAP) is a method for reducing the dimensionality of a data set (
Becht E et al. (2018))
Gene and protein expression levels for different datasets are expressed as transcripts per million ("TPM"), protein-transcripts per million ("pTPM") and normalized expression ("nTPM").
The default content is a
Association containing a the expression levels (nTPM) of genes in different human tissues along with these additional data:
| "TissueAtlas Gene co-expression network" | list of gene pairs with high correlation (>0.7) in expression in tissues |
| "TissueAtlas Gene co-expression graph" | graph of co-expressing genes in tissues |
| "TissueAtlas Gene co-expression network Graph measures" | graph measures for the "TissueAtlas Gene co-expression network" |
| "TissueAtlas maximum expression location" | maximum expression location of genes |
| "BrainAtlas gene expression (TPM)" | expression levels (TPM) of genes in human brain |
| "BrainAtlas gene expression (pTPM)" | expression levels (pTPM) of genes in human brain |
| "BrainAtlas gene expression (nTPM)" | expression levels (nTPM) of genes in human brain |
| "BrainAtlas Gene co-expression network" | list of gene pairs with high correlation (>0.7) in expression in human brain |
| "BrainAtlas Gene co-expression graph" | graph of co-expressing genes in human brain |
| "BrainAtlas Gene co-expression network Graph measures" | graph measures for the "BrainAtlas Gene co-expression network" |
| "PathologyAtlas" | data about roles of genes in different cancers |
| "PathologyAtlas mRNA expression (FPKM) "<>cancer | gene expression (FPKM) values for different genes in different cancer types |
| "PathologyAtlas Samples" | sample details for gene expression data in different cancers |
| "SingleCellAtlas expression (nTPM)" | expression levels (nTPM) of genes in different cell types |
| "SingleCellAtlas cell clusters" | description of cell clusters |
| "SingleCellAtlas expression in cell clusters(nTPM)" | expression levels (nTPM) of genes in different cell types and clusters |
| "SingleCellAtlas Gene co-expression network" | list of gene pairs with high correlation (>0.7) in expression in single cell types |
| "SingleCellAtlas Gene co-expression graph" | graph of co-expressing genes in different human single cell types |
| "SingleCellAtlas Gene co-expression network Graph measures" | graph measures for the "SingleCellAtlas Gene co-expression network" |
| "SingleCellAtlas UMAP coordinates in tissue "<>tissue | UMAP coordinates for cells in clusters for different tissues |
| "SubCellularAtlas" | expression of genes in different subcellular regions |
| "Ensembl ID gene name association" | Association of Emsembl IDs of genes and common gene names |
| "Ensembl ID gene description UniProtID association" | Association of Emsembl IDs of genes to gene description and UniProtID |
| "Organ tissue association" | Association of organs and tissues belonging to an organ |
"PathologyAtlas mRNA expression (FPKM) "<>cancer has the gene expression (FPKM) values for different genes in different cancer types for different samples in The Cancer Genome Atlas.
cancer has the following values: "BRCA","BLCA","CESC","COAD","READ","GBM","HNSC","KIRC","KIRP","KICH","LIHC","LUAD","LUSC","OV","PAAD","PRAD","SKCM","STAD","TGCT","THCA","UCEC". These are abbreviations for different cancer types
tissue for UMAP coordinates can be adipose_tissue, bone_marrow, brain, breast, bronchus, colon, endometrium, esophagus, eye, fallopian_tube, heart_muscle, kidney, liver, lung, lymph_node, ovary, pancreas, pbmc, placenta, prostate, rectum, salivary_gland, skeletal_muscle, skin, small_intestine, spleen, stomach, testis, thymus, tongue and vascular.

![RNAExpressionData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\)];
tissues = RNAExpressionData[[1]];
ensemblIDGeneAssoc = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "Ensembl ID gene name association"];
ensemblIDs = Keys@RNAExpressionData[[2 ;; -1]];
genes = ensemblIDGeneAssoc /@ ensemblIDs;
v = Values@RNAExpressionData;
totExp = Table[Total@v[[i]] /. {"NA" -> 0}, {i, 2, Length@genes + 1}];
sv = Sort[totExp];
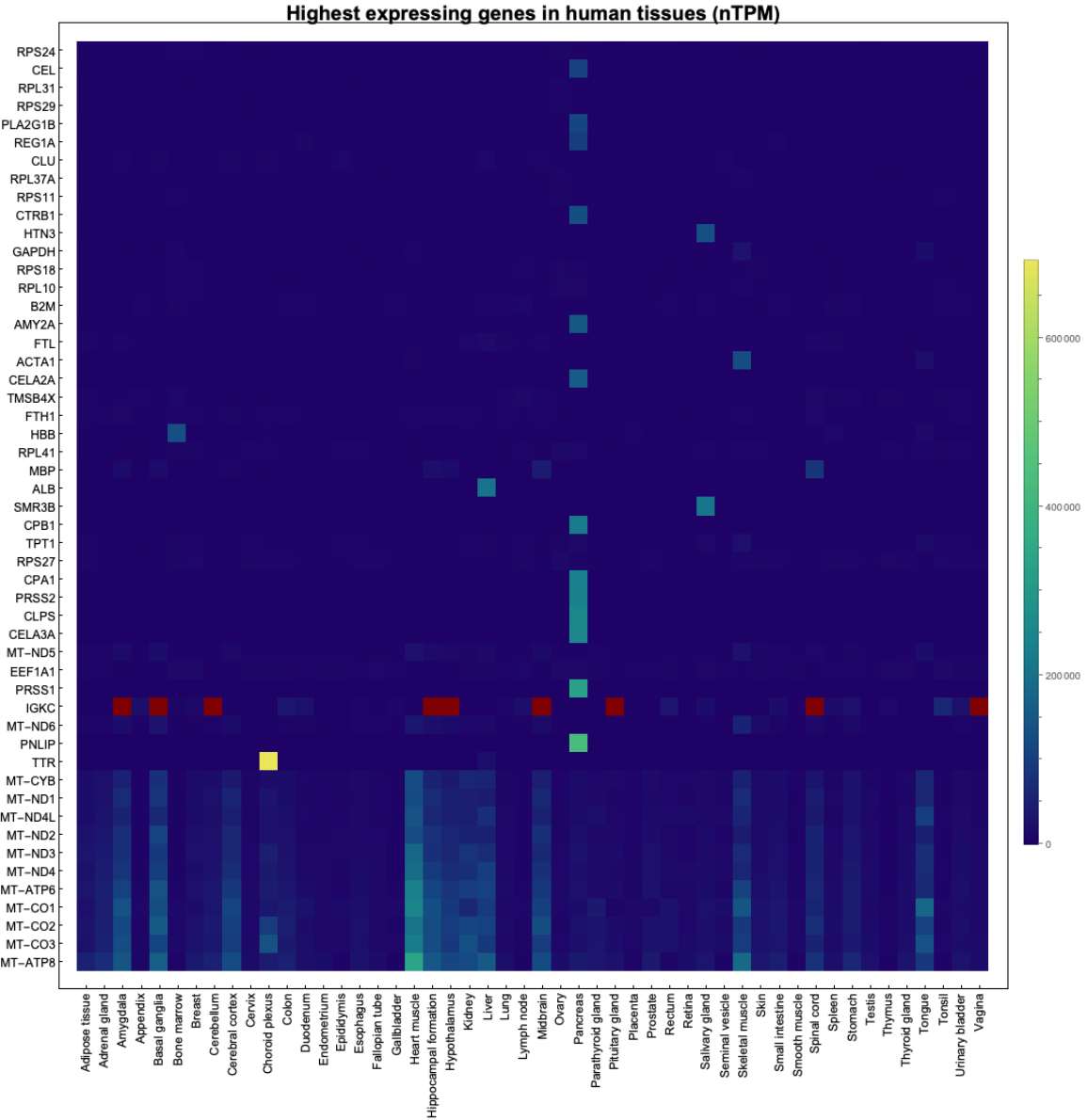
numGenes = 50;
pos = Flatten@
Map[Position[totExp, #] &, sv[[Length@genes - numGenes ;; Length@genes]]];
highestExpressionGenes = ensemblIDs[[pos]];
highestExpressionGeneNames = Map[ensemblIDGeneAssoc, highestExpressionGenes];
ftx = MapThread[{#1, #2} &, {Range@Length@highestExpressionGeneNames, highestExpressionGeneNames}];
fty = MapThread[{#1, Rotate[Capitalize@#2, Pi/2]} &, {Range@
Length@tissues, tissues}];
array = v[[pos + 1]];
mA = {Min@array /. "NA" -> 0, Max@array /. "NA" -> 0};
ArrayPlot[array, ColorFunction -> "BlueGreenYellow", FrameTicks -> {ftx, fty}, ImageSize -> 1000, Frame -> True, FrameStyle -> Directive[Black, 12], PlotLegends -> BarLegend[{"BlueGreenYellow", mA}], PlotLabel -> Style["Highest expressing genes in human tissues (nTPM)", Black, Bold, 20]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/1e0129babf5be351.png)
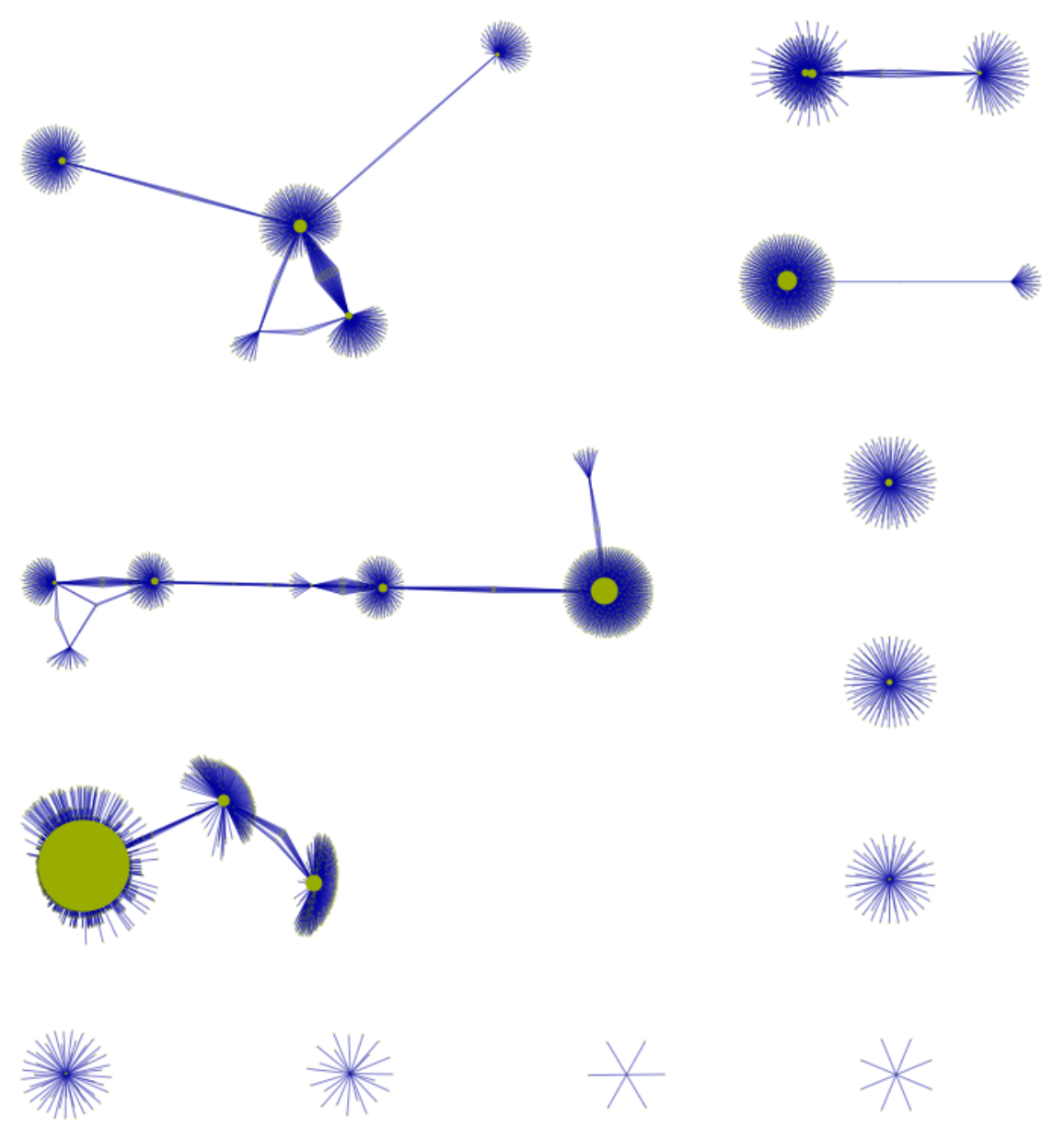
![RNACoExpressionNetwork = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "TissueAtlas Gene co-expression graph"];
getSubGraph[graph_Graph, vertexList_] := Graph@Sort@
DeleteDuplicates@
Flatten[IncidenceList[graph, #] & /@ vertexList]; ResourceFunction[
ResourceObject[<|"Name" -> "VertexSizeScaledGraph", "ShortName" -> "VertexSizeScaledGraph", "UUID" -> "82dfde6e-106f-4322-a329-1216e9f28362", "ResourceType" -> "Function", "Version" -> "1.1.0", "Description" -> "Visualize a graph with scaled vertex size based on custom graph related measures", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$4d8c3d76bc4e42be9489e98149696195`VertexSizeScaledGraph", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/b0eb1e52-6cf3-4b6a-8efb-7950a9c63da5"]|>, ResourceSystemBase -> Automatic]][
getSubGraph[RNACoExpressionNetwork, Echo@RandomChoice[VertexList@RNACoExpressionNetwork, 30]], VertexStyle -> RGBColor[0.6, 0.68, 0], EdgeStyle -> RGBColor[0, 0, 0.61], "MaxVertexSize" -> 200, VertexLabels -> Placed["Name", Tooltip]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/555e7cb240b060d0.png)

![RNAExpressionData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\)];
RNACoExpressionNetwork = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "TissueAtlas Gene co-expression network"];
ensemblIDGeneAssoc = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "Ensembl ID gene name association"];
ensemblIDs = Keys@RNAExpressionData[[2 ;; -1]];
genes = ensemblIDGeneAssoc /@ ensemblIDs;
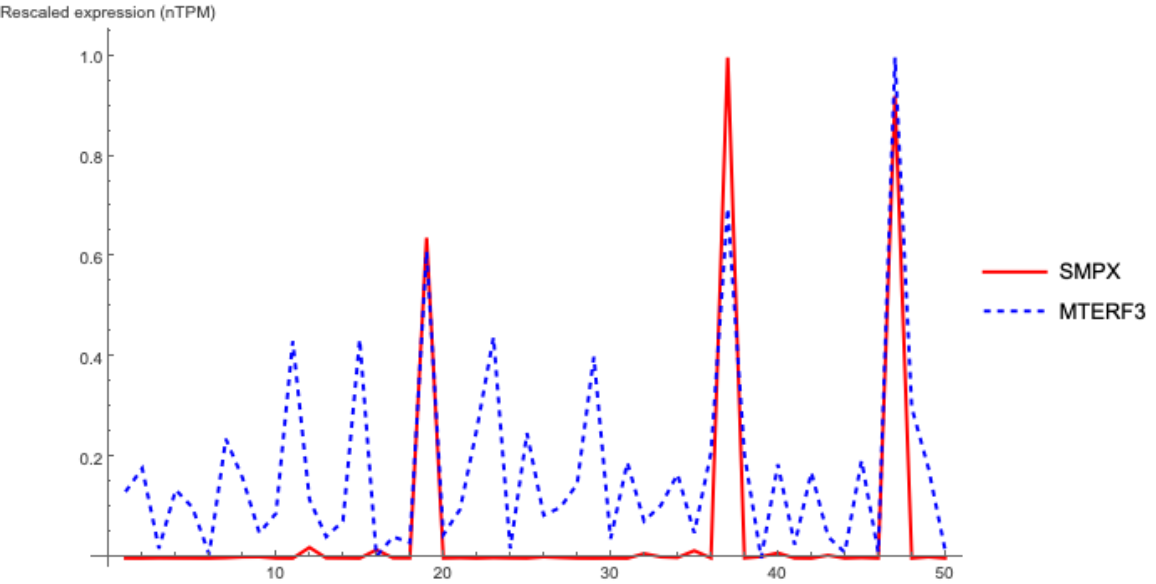
genePairs = VertexList[Graph@RandomChoice@EdgeList@RNACoExpressionNetwork];
array = (Rescale@RNAExpressionData[#] &) /@ ensemblIDs[[Flatten[Position[genes, #] & /@ genePairs]]];
ListLinePlot[array, PlotLegends -> {genePairs[[1]], genePairs[[2]]}, AxesLabel -> {None, "Rescaled expression (nTPM)" }, PlotRange -> All,
PlotStyle -> {Red, Directive[Blue, Dashed]}, ImageSize -> Large]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/5a0f8ceabfba1e72.png)
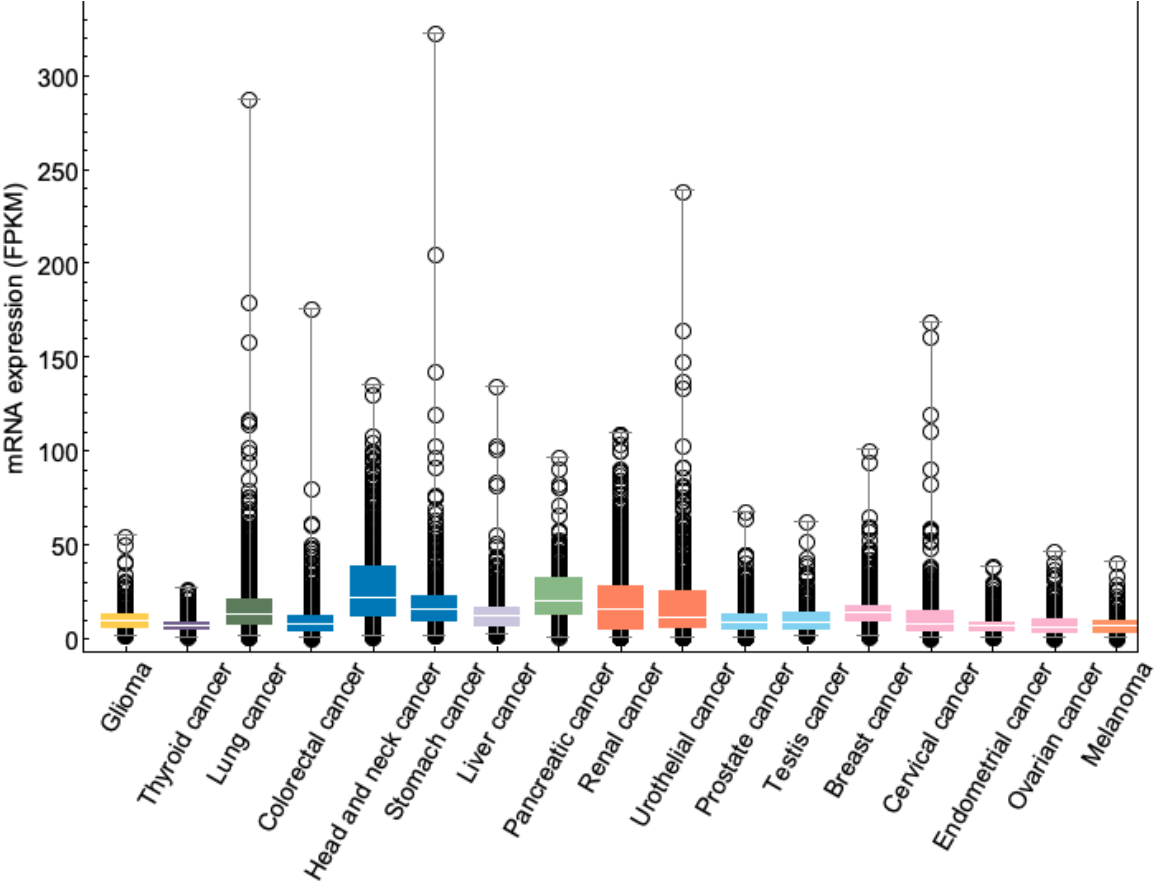
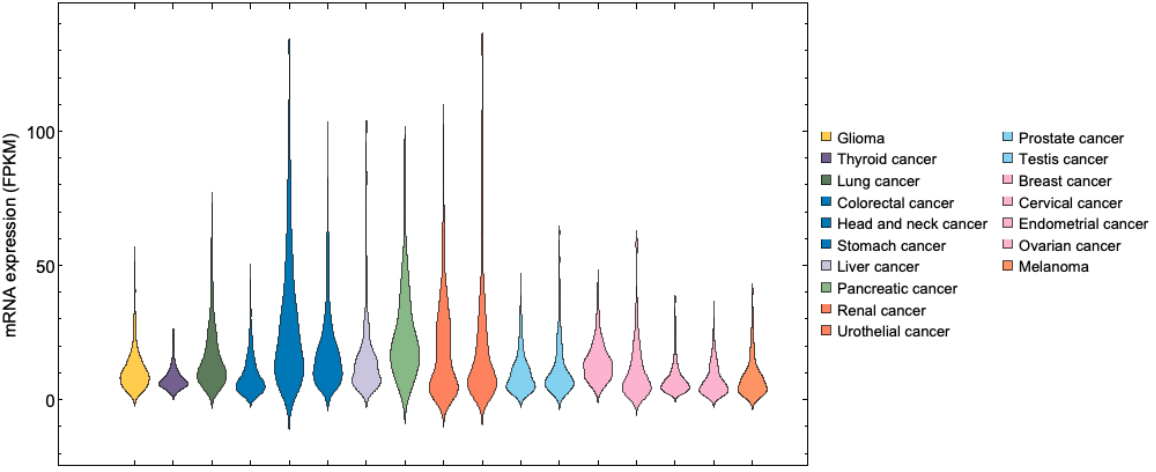
![plotCancerShortNames = {{"GBM"}, {"THCA"}, {"LUAD", "LUSC"}, {"COAD", "READ"}, {"HNSC"}, {"STAD"}, {"LIHC"}, {"PAAD"}, {"KIRC", "KIRP", "KICH"}, {"BLCA"}, {"PRAD"}, {"TGCT"}, {"BRCA"}, {"CESC"}, {"UCEC"}, {"OV"}, {"SKCM"}};
plotCancerLongNames = {"Glioma", "Thyroid cancer", "Lung cancer", "Colorectal cancer", "Head and neck cancer", "Stomach cancer", "Liver cancer", "Pancreatic cancer", "Renal cancer", "Urothelial cancer", "Prostate cancer", "Testis cancer", "Breast cancer", "Cervical cancer", "Endometrial cancer", "Ovarian cancer", "Melanoma"};
plotCancerColors = {
RGBColor[1., 0.800000094944996, 0.3019609068561773, 1.],
RGBColor[
0.454901902606013, 0.37647054361669646`, 0.5568627171197533, 1.],
RGBColor[
0.35816992186625884`, 0.48104578647795015`, 0.35294121107541104`, 1.],
RGBColor[
9.589812169208401*^-7, 0.4705881904500226, 0.7176470465066893, 1.],
RGBColor[
9.589812169208401*^-7, 0.4705881904500226, 0.7176470465066893, 1.],
RGBColor[
9.589812169208401*^-7, 0.4705881904500226, 0.7176470465066893, 1.],
RGBColor[
0.7960782989163758, 0.7686276968328074, 0.8745099497269464, 1.],
RGBColor[
0.5372548827993883, 0.7215686797169252, 0.5294118166131165, 1.],
RGBColor[1., 0.509803856871976, 0.3725490654639468, 1.],
RGBColor[1., 0.509803856871976, 0.3725490654639468, 1.],
RGBColor[
0.5137255717824479, 0.8196078836750866, 0.941176564576014, 1.],
RGBColor[
0.5137255717824479, 0.8196078836750866, 0.941176564576014, 1.],
RGBColor[
0.9832875177291199, 0.7011909727286483, 0.8148310349772592, 1.],
RGBColor[
0.9832875177291199, 0.7011909727286483, 0.8148310349772592, 1.],
RGBColor[
0.9832875177291199, 0.7011909727286483, 0.8148310349772592, 1.],
RGBColor[
0.9832875177291199, 0.7011909727286483, 0.8148310349772592, 1.],
RGBColor[
0.9946701221664577, 0.575625452943115, 0.3876721337241307, 1.]};
cancers = {"BRCA", "BLCA", "CESC", "COAD", "READ", "GBM", "HNSC", "KIRC", "KIRP", "KICH", "LIHC", "LUAD", "LUSC", "OV", "PAAD", "PRAD", "SKCM", "STAD", "TGCT", "THCA", "UCEC"};
pathologyData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "PathologyAtlas mRNA expression (FPKM) " <> #] & /@ cancers;
dataFull = AssociationThread[cancers, pathologyData];](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/6a4700747de846d9.png)
![plotCancerColorsAssoc = AssociationThread[plotCancerLongNames, plotCancerColors];
plotCancerTickLabelsAssoc = AssociationThread[plotCancerLongNames, plotCancerShortNames];
ensGeneAssoc = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "Ensembl ID gene name association"];
geneEnsAssoc = AssociationThread[Values@ensGeneAssoc, Keys@ensGeneAssoc];
ticks = MapThread[{#1, Rotate[#2, -5*Pi/3]} &, {Range@
Length@plotCancerLongNames, plotCancerLongNames}];
cancerGeneExpressionPlot[gene_] := Module[{ensembl, data, plottingData, xVal, yVals, plotData}, ensembl = geneEnsAssoc@gene;
data = (#@ensembl) & /@ dataFull;
plottingData = Table[xVal = Position[plotCancerLongNames, cancerNameToPlot][[1, 1]];
yVals = Flatten[data[#] & /@ plotCancerTickLabelsAssoc[cancerNameToPlot]];
plotData = {xVal, #} & /@ yVals;
{plotData, yVals}, {cancerNameToPlot, plotCancerLongNames}];
Show[ListPlot[plottingData[[All, 1]], PlotStyle -> Black, PlotMarkers -> "\!\(\*
GraphicsBox[CircleBox[{0, 0}],
ImageSize->{11., Automatic}]\)", FrameTicks -> {{Automatic, None}, {ticks, None}}, PlotRange -> Full, ImageSize -> 700, Frame -> True, FrameLabel -> {None, "mRNA expression (FPKM)"}, FrameStyle -> Directive[Black, 15]], BoxWhiskerChart[plottingData[[All, 2]], ChartStyle -> plotCancerColors]]]
cancerGeneExpressionDistributionChart[gene_] := Module[{ensembl, data, plottingData}, ensembl = geneEnsAssoc@gene;
data = (#@ensembl) & /@ dataFull;
plottingData = Table[xVal = Position[plotCancerLongNames, cancerNameToPlot][[1, 1]];
Flatten[
data[#] & /@ plotCancerTickLabelsAssoc[cancerNameToPlot]], {cancerNameToPlot,
plotCancerLongNames}];
DistributionChart[plottingData, ChartStyle -> plotCancerColors, ChartLegends -> plotCancerLongNames, PlotRange -> Automatic, ImageSize -> 700, FrameLabel -> {None, "mRNA expression (FPKM)"}, FrameStyle -> Directive[Black, 15]]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/24d8cb6a9bdcdbfa.png)
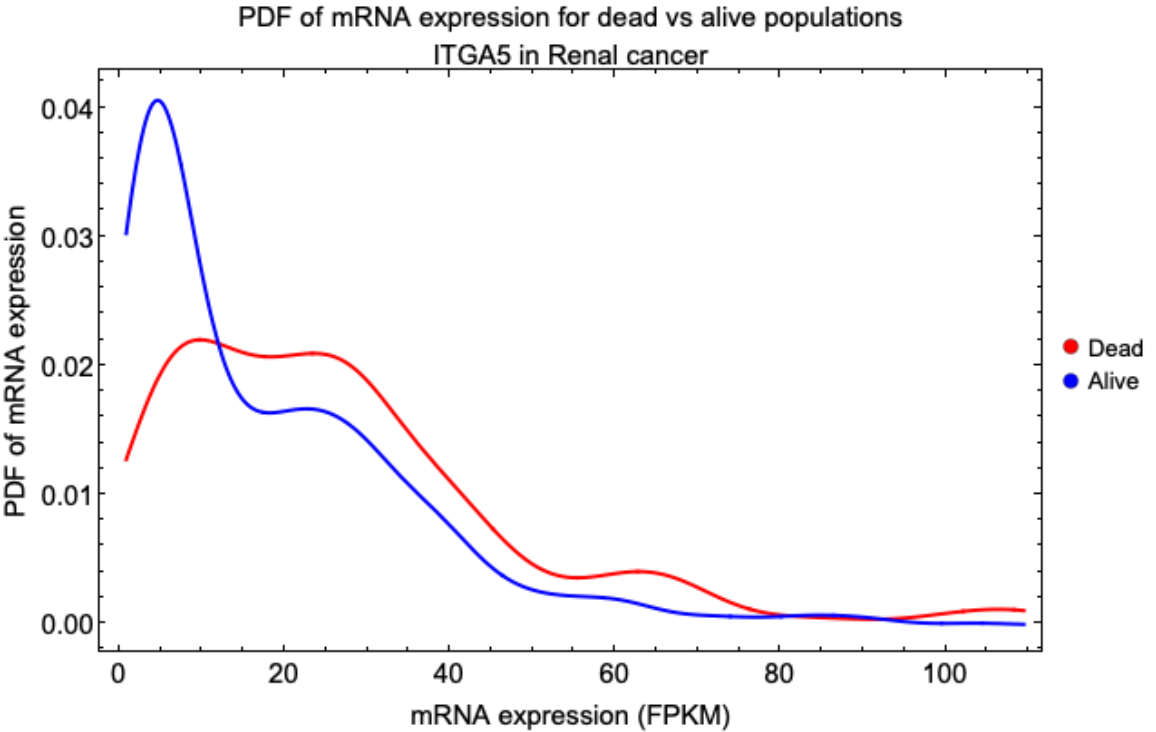
![(* Evaluate this cell to get the example input *) CloudGet["https://www.wolframcloud.com/obj/9a3d4ec7-24c2-4d03-90d7-6ac26111a2da"]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/475bd3c698d926d8.png)

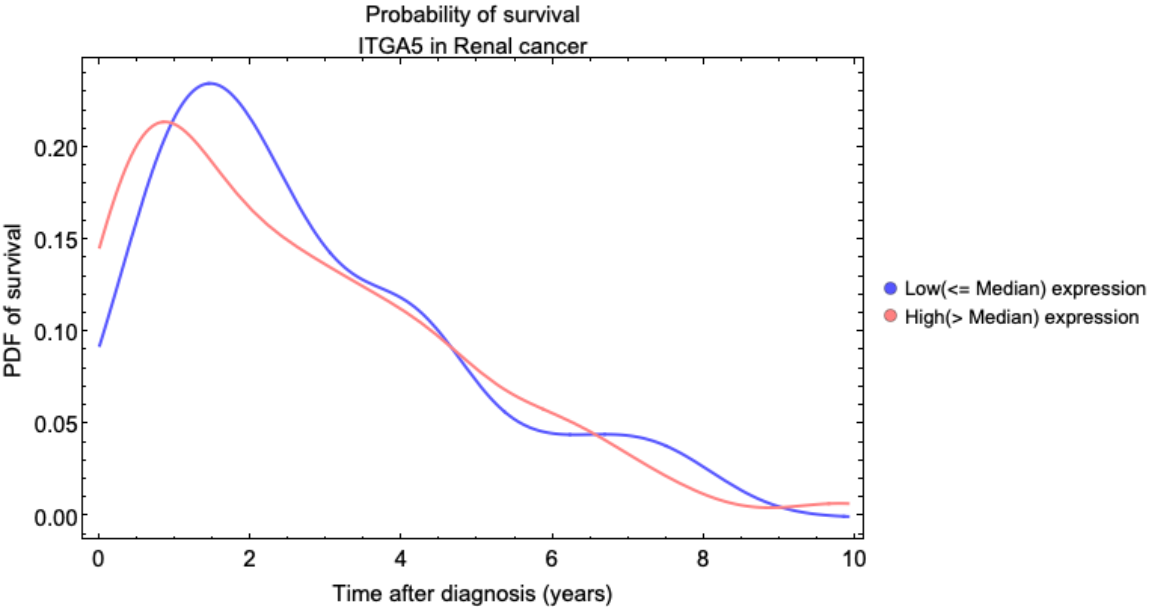
![CellClusterPlot[organ_] := Module[{assoc, clusters, L, color, clusterList},
assoc = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "SingleCellAtlas UMAP coordinates in tissue " <> organ];
clusters = Keys@assoc;
clusterList = StringJoin["Cluster ", ToString@#] & /@ clusters;
L = Length@clusters;
color = ColorData["Rainbow"][#] & /@ (Range[L]/L);
Legended[
ListPlot[Table[Style[assoc[[i, All, -1]], color[[i]]], {i, L}], Axes -> False, FrameLabel -> {"UMAP_x", "UMAP_y"}, FrameStyle -> Directive[Black, Thickness[0.0025], 15], AspectRatio -> 1, PlotLabel -> Style[Capitalize@organ, Black, 18],
Frame -> True, PlotStyle -> PointSize[Small]], SwatchLegend[color, clusterList, LegendMarkers -> "Bubble"]]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/3cc23aa18a056f2a.png)
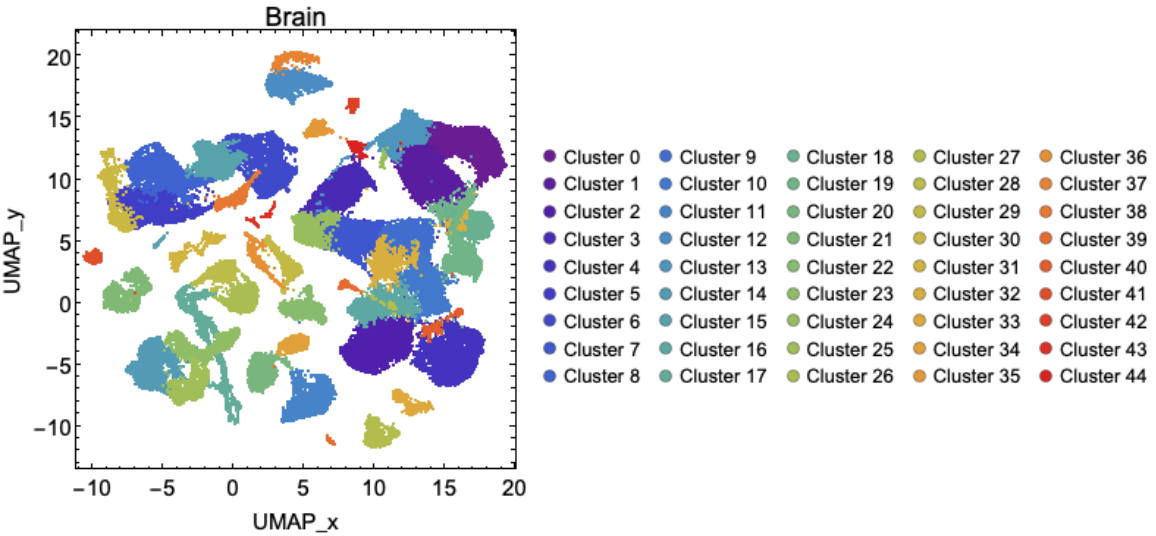
![CellClusterPlotBasic[organ_] := Module[{assoc, clusters, L, color, clusterList},
assoc = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "SingleCellAtlas UMAP coordinates in tissue " <> organ];
clusters = Keys@assoc;
clusterList = StringJoin["Cluster ", ToString@#] & /@ clusters;
L = Length@clusters;
color = ColorData["Rainbow"][#] & /@ (Range[L]/L);
ListPlot[Table[Style[assoc[[i, All, -1]], color[[i]]], {i, L}], Axes -> False, FrameLabel -> {"UMAP_x", "UMAP_y"}, FrameStyle -> Directive[Black, Thickness[0.0025], 15], AspectRatio -> 1, PlotLabel -> Style[Capitalize@organ, Black, 18], Frame -> True, PlotStyle -> PointSize[Small]]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/342439d6a05d167f.png)

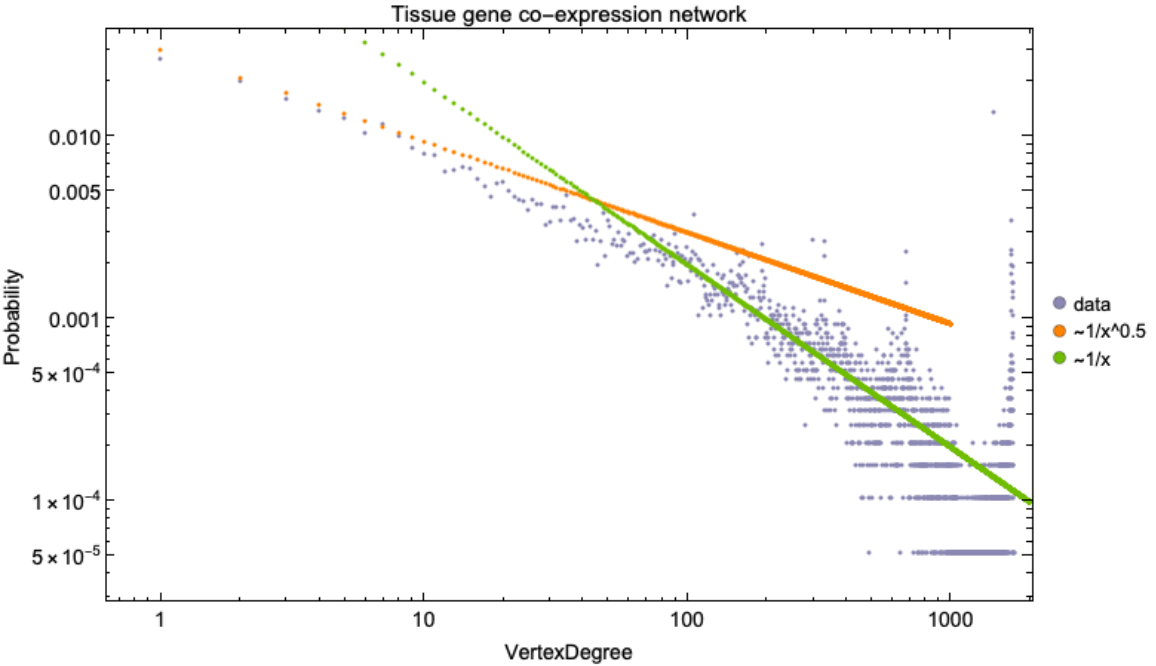
![RNACoExpressionNetwork = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "TissueAtlas Gene co-expression graph"];
power = 1/2;
power2 = 1;
Legended[Show[ListLogLogPlot[ResourceFunction[
ResourceObject[<|"Name" -> "VertexDegreeRelativeFrequencies", "ShortName" -> "VertexDegreeRelativeFrequencies", "UUID" -> "a76d6242-2a31-4326-bb6a-df4e446f50a6", "ResourceType" -> "Function", "Version" -> "1.0.0", "Description" -> "Relative frequencies of VertexDegree in a Graph", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$8f984335e1b840a0b3c49438547b6dc4`VertexDegreeRelativeFrequencies", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/beb446d6-a18b-4eae-b8fa-c7d76afd70c6"]|>, ResourceSystemBase -> Automatic]][
RNACoExpressionNetwork], ImageSize -> 700, PlotStyle -> RGBColor[0.54, 0.53, 0.7000000000000001], Frame -> True, FrameStyle -> Directive[Black, 14], PlotLabel -> Style["Tissue gene co-expression network", Black, 15],
FrameLabel -> {{"Probability", None}, {"VertexDegree", Automatic}}],
ListLogLogPlot[(0.03/#^power) & /@ (Range@1000), PlotStyle -> RGBColor[1, 0.51, 0]], ListLogLogPlot[(0.2/#^power2) & /@ (Range@2000), PlotStyle -> RGBColor[0.43, 0.74, 0]]], SwatchLegend[{RGBColor[0.54, 0.53, 0.7000000000000001], RGBColor[
1, 0.51, 0], RGBColor[0.43, 0.74, 0]}, {"data", "~1/x^0.5", "~1/x"},
LegendMarkers -> "Bubble"]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/34bfeabb90c3ddc8.png)
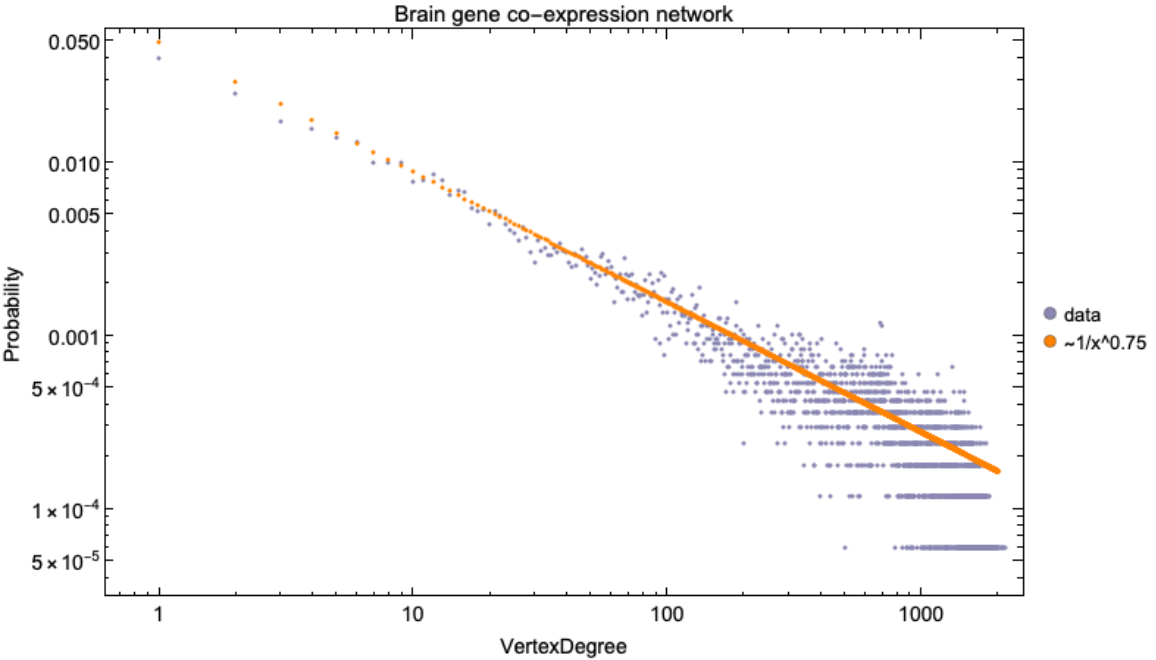
![brainGeneCoExpressionNetwork = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "BrainAtlas Gene co-expression graph"];
\[Alpha] = 3/4;
Legended[Show[ListLogLogPlot[ResourceFunction[
ResourceObject[<|"Name" -> "VertexDegreeRelativeFrequencies", "ShortName" -> "VertexDegreeRelativeFrequencies", "UUID" -> "a76d6242-2a31-4326-bb6a-df4e446f50a6", "ResourceType" -> "Function", "Version" -> "1.0.0", "Description" -> "Relative frequencies of VertexDegree in a Graph", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$8f984335e1b840a0b3c49438547b6dc4`VertexDegreeRelativeFrequencies", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/beb446d6-a18b-4eae-b8fa-c7d76afd70c6"]|>, ResourceSystemBase -> Automatic]][
brainGeneCoExpressionNetwork], ImageSize -> 700, PlotStyle -> RGBColor[0.54, 0.53, 0.7000000000000001], Frame -> True, FrameStyle -> Directive[Black, 14], FrameLabel -> {{"Probability", None}, {"VertexDegree", Automatic}},
PlotLabel -> Style["Brain gene co-expression network", Black, 15]],
ListLogLogPlot[(0.05/#^\[Alpha]) & /@ (Range@2000), PlotStyle -> RGBColor[1, 0.51, 0]]], SwatchLegend[{RGBColor[0.54, 0.53, 0.7000000000000001], RGBColor[
1, 0.51, 0]}, {"data", "~1/x^0.75"}, LegendMarkers -> "Bubble"]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/21411fb41fd2a953.png)
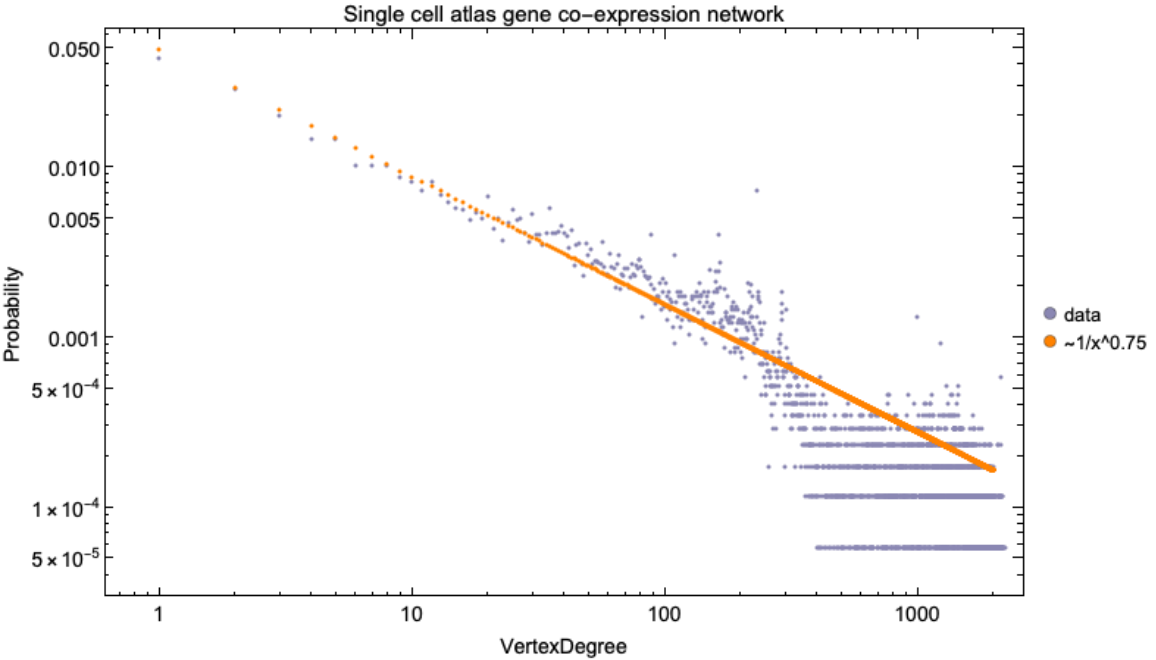
![scGeneCoExpressionNetwork = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "SingleCellAtlas Gene co-expression graph"];
\[Alpha] = 3/4;
Legended[Show[ListLogLogPlot[ResourceFunction[
ResourceObject[<|"Name" -> "VertexDegreeRelativeFrequencies", "ShortName" -> "VertexDegreeRelativeFrequencies", "UUID" -> "a76d6242-2a31-4326-bb6a-df4e446f50a6", "ResourceType" -> "Function", "Version" -> "1.0.0", "Description" -> "Relative frequencies of VertexDegree in a Graph", "RepositoryLocation" -> URL[
"https://www.wolframcloud.com/obj/resourcesystem/api/1.0"], "SymbolName" -> "FunctionRepository`$8f984335e1b840a0b3c49438547b6dc4`VertexDegreeRelativeFrequencies", "FunctionLocation" -> CloudObject[
"https://www.wolframcloud.com/obj/beb446d6-a18b-4eae-b8fa-c7d76afd70c6"]|>, ResourceSystemBase -> Automatic]][
scGeneCoExpressionNetwork], ImageSize -> 700, PlotStyle -> RGBColor[0.54, 0.53, 0.7000000000000001], Frame -> True, FrameStyle -> Directive[Black, 14], FrameLabel -> {{"Probability", None}, {"VertexDegree", Automatic}},
PlotLabel -> Style["Single cell atlas gene co-expression network", Black, 15]],
ListLogLogPlot[(0.05/#^\[Alpha]) & /@ (Range@2000), PlotStyle -> RGBColor[1, 0.51, 0]]], SwatchLegend[{RGBColor[0.54, 0.53, 0.7000000000000001], RGBColor[
1, 0.51, 0]}, {"data", "~1/x^0.75"}, LegendMarkers -> "Bubble"]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/4881426eb0dd2cb0.png)
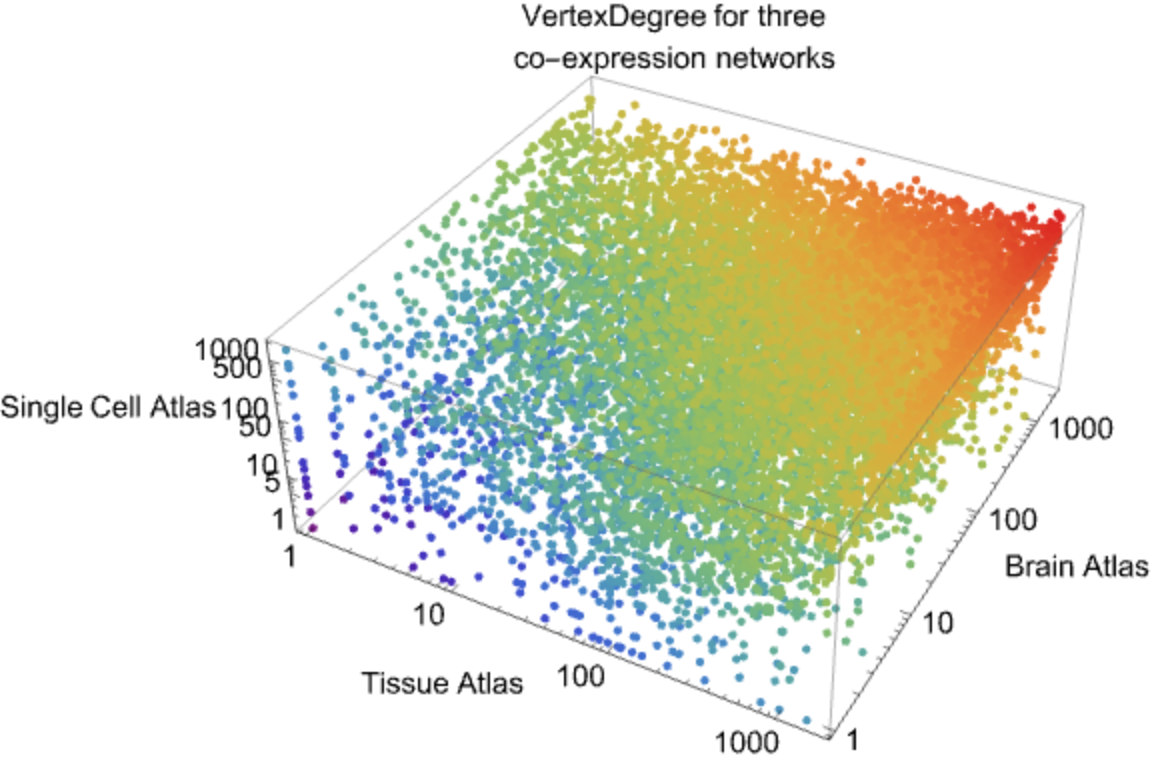
![commonGenes = Intersection[VertexList@ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "TissueAtlas Gene co-expression graph"], VertexList@ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "BrainAtlas Gene co-expression graph"], VertexList@ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "SingleCellAtlas Gene co-expression graph"]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/1c57d2200b46bbbb.png)

![tissueData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "TissueAtlas Gene co-expression network Graph measures"];
brainData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "BrainAtlas Gene co-expression network Graph measures"];
scData = ResourceData[\!\(\*
TagBox["\"\<Human Protein Atlas\>\"",
#& ,
BoxID -> "ResourceTag-Human Protein Atlas-Input",
AutoDelete->True]\), "SingleCellAtlas Gene co-expression network Graph measures"];
maxV = Max@
MapThread[#1* #2* #3 &, {Map[tissueData[#, "VertexDegree"] &, commonGenes], Map[brainData[#, "VertexDegree"] &, commonGenes], Map[scData[#, "VertexDegree"] &, commonGenes]}];
plotVDData = MapThread[
Tooltip[Style[{#1, #2, #3}, ColorData["Rainbow"][Log[#1* #2* #3]/Log@maxV]], #4] &, {Map[
tissueData[#, "VertexDegree"] &, commonGenes], Map[brainData[#, "VertexDegree"] &, commonGenes], Map[scData[#, "VertexDegree"] &, commonGenes], commonGenes}];](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/487222768ca6350a.png)
![ListPointPlot3D[plotVDData, ScalingFunctions -> {"Log", "Log", "Log"},
PlotStyle -> PointSize[Medium], AxesStyle -> Directive[Black, 15], AxesLabel -> {"Tissue Atlas", "Brain Atlas", "Single Cell Atlas"}, ImageSize -> Large, PlotLabel -> Style["VertexDegree for three\nco-expression networks", Black, 15]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/4925213b64335967.png)
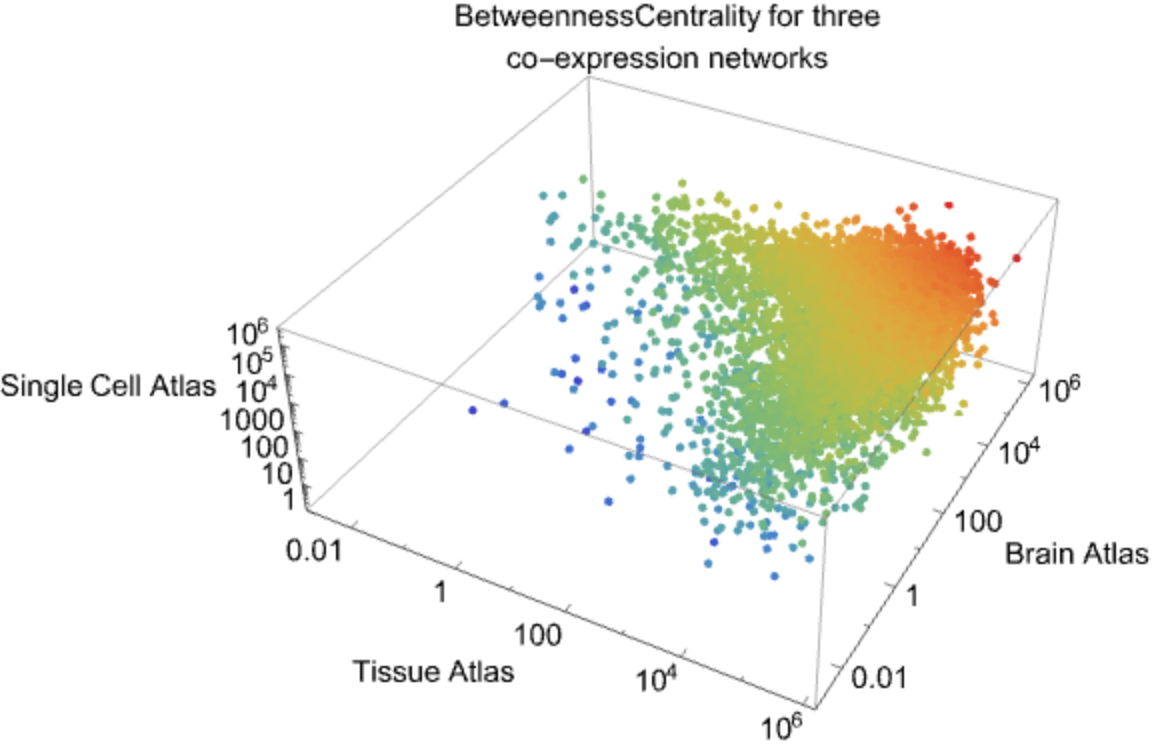
![maxV = Max@
MapThread[#1* #2* #3 &, {Map[
tissueData[#, "BetweennessCentrality"] &, commonGenes], Map[brainData[#, "BetweennessCentrality"] &, commonGenes], Map[scData[#, "BetweennessCentrality"] &, commonGenes]}];
plotBCData = MapThread[
Tooltip[Style[{#1, #2, #3}, ColorData["Rainbow"][
Log[If[#1* #2* #3 == 0, 0.000001, #1* #2* #3]]/
Log@maxV]], #4] &, {Map[
tissueData[#, "BetweennessCentrality"] &, commonGenes], Map[brainData[#, "BetweennessCentrality"] &, commonGenes], Map[scData[#, "BetweennessCentrality"] &, commonGenes], commonGenes}];
ListPointPlot3D[plotBCData, ScalingFunctions -> {"Log", "Log", "Log"},
PlotStyle -> PointSize[Medium], AxesStyle -> Directive[Black, 15], AxesLabel -> {"Tissue Atlas", "Brain Atlas", "Single Cell Atlas"}, ImageSize -> Large, PlotLabel -> Style["BetweennessCentrality for three\nco-expression networks", Black, 15]]](https://www.wolframcloud.com/obj/resourcesystem/images/d9f/d9f76eb9-4537-4037-9e88-d054bd89b2b1/094634c7e0464ee9.png)