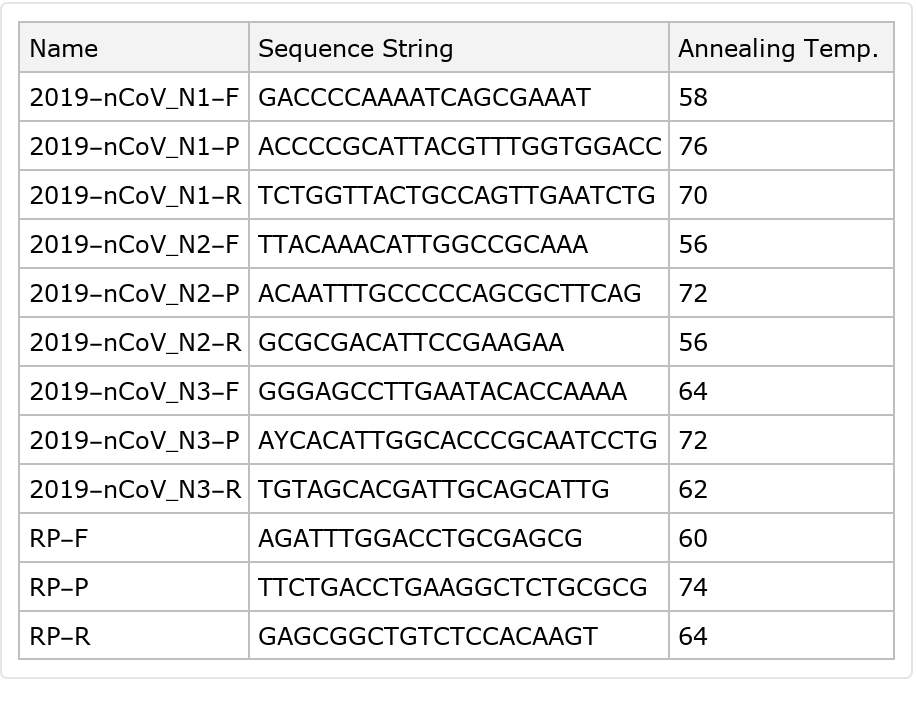
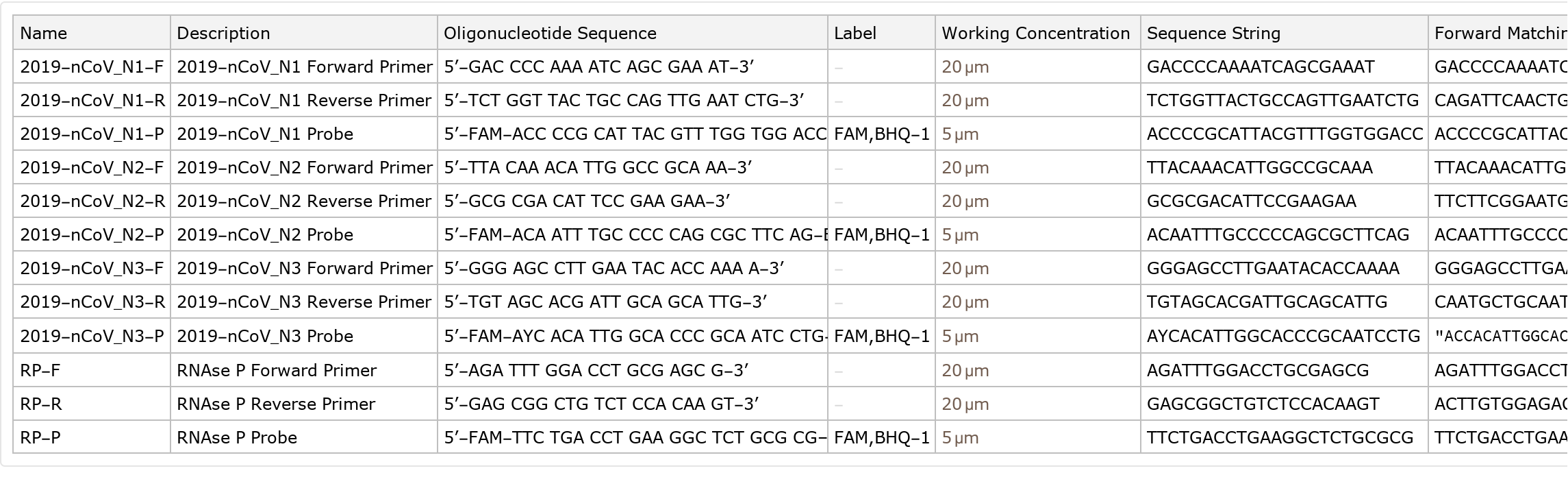
Primers provided by the US Centers for Disease Control and Prevention (CDC) for identifying SARS-CoV-2 for research purposes, including the names, sequences, working concentration, and related information
Examples
Basic Examples
Observe the different concentrations applied by each label:
Visualizations
With a few exceptions, the CDC primers designed for matching SARS-CoV-2 sequences do match the complete sequence as intended, either in translated order or from the reverse complement, as designated. The RNAse P primers are not for matching SARS-CoV-2, but for making sure that there are appropriate background levels of human protein for the test to be considered valid, and so are designed not to match:
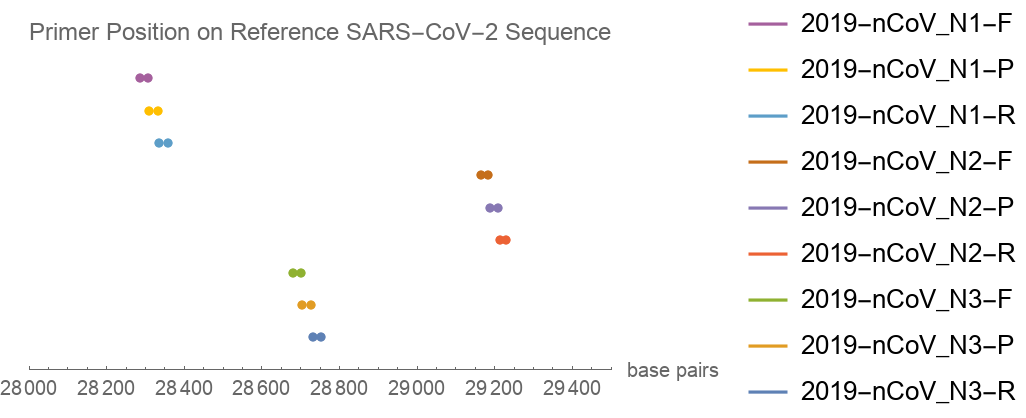
The primers target a fairly narrow range of the viral genome, which is as designed:
Analysis
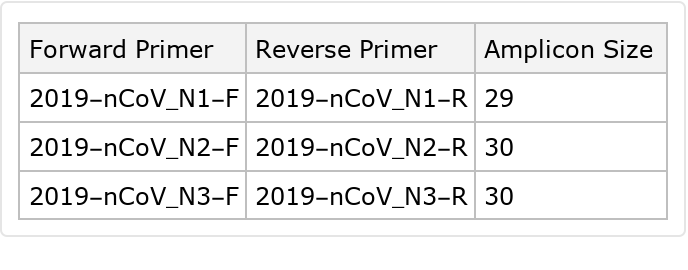
PCR experiments typically aim for the size of the genetic materials to be amplified for measurement (the amplicon) to be around 1000 base pairs and hopefully smaller for a high product yield:
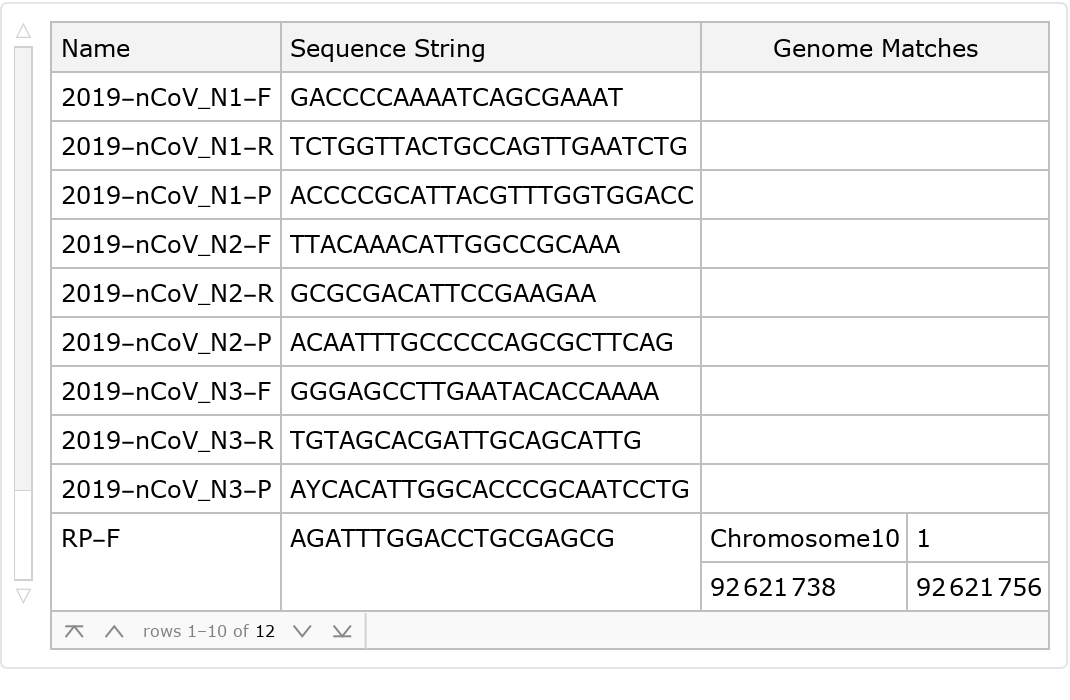
A primer aimed at matching a virus should not also match sequences within the target genome. Here, we see that matches are found only for the RP primers aimed at establishing a significant background of human genetic material in the sample, as desired:
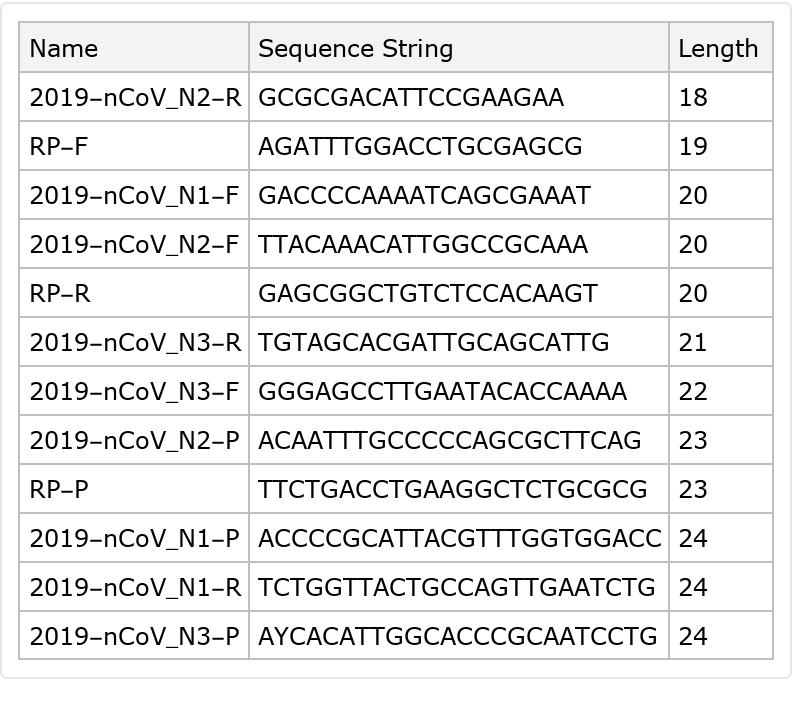
An ideal length for primers is around 18 to 22 base pairs long. Primer length can affect specificity and yield; the longer the primers, the more specific they are (i.e. the less likely they will anneal to sequences you did not design them for), but the less often they anneal at all (to correct sequences or otherwise):
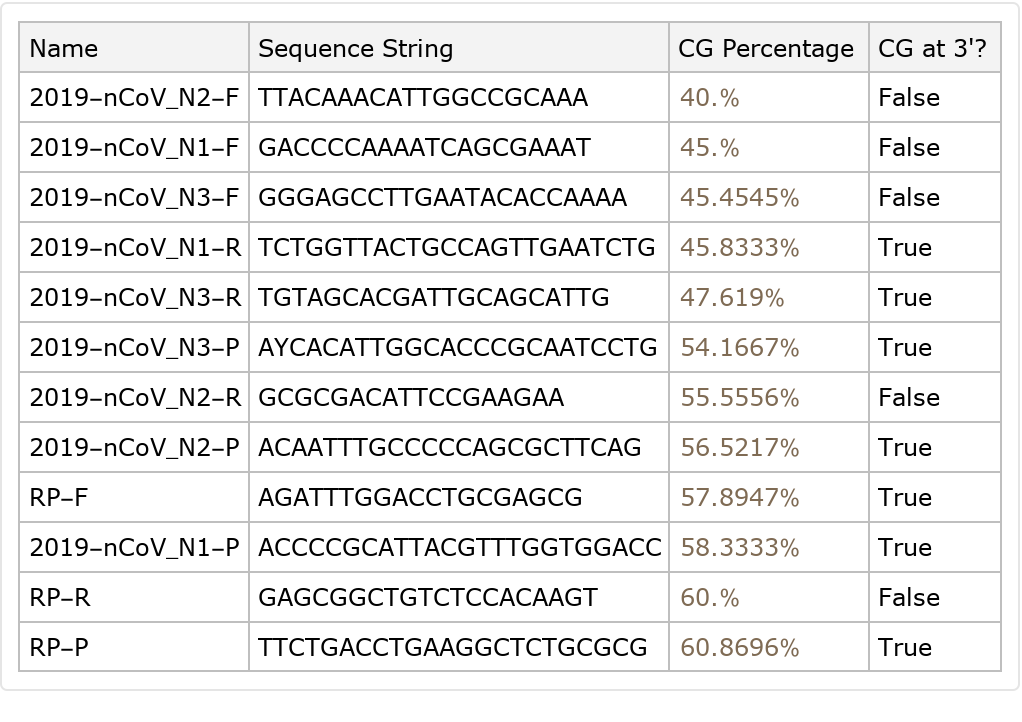
One factor in making a good primer is the percentage of cytosine and guanine the primer is composed of . A CG content of 40%-60% is generally accepted as producing the best results, while a C or G at the 3' end of the primer can promote binding specificity:
The length and GC content of primers can affect melting temperature (Tm) of the primers, as longer primers and higher GC content will mean the primer requires more energy, and a higher temperature, to anneal. Primers within a common temperature band will have the best signal-to-noise ratio in their detection results. Primers should usually be within 5˚C of difference in Tm, so it would probably be best to pay close attention to which of these primers are used together:
Bibliographic Citation
Wolfram Research,
"CDC Primers for SARS-CoV-2 Research"
from the Wolfram Data Repository
(2020)
These sequences are intended to be used for the purposes of respiratory virus surveillance and research. The recipient agrees to use them in compliance with all applicable laws and regulations. Every effort has been made to assure the accuracy of the sequences, but CDC cannot provide any warranty regarding their accuracy. The recipient can acknowledge the source of sequences in any oral presentations or written publications concerning the research project by referring to the Division of Viral Diseases, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA.
Data Resource History
See Also
Publisher Information

![accessionToViralSequence = <|
Rule @@@ (Normal[
Values /@ Select[ResourceData[
"Genetic Sequences for the SARS-CoV-2 Coronavirus"], StringContainsQ[#GenBankTitle, "complete genome"] &][[
All, {"Accession", "Sequence"}]]])|>;
primerToForwardPattern = <|
Rule @@@ Normal[Values /@ ResourceData["CDC Primers for SARS-CoV-2 Research"][[
All, {"Name", "Forward Matching Pattern"}]]]|>;
listToTickLabels[lst_] := MapIndexed[{#2[[1]], #1} &, lst];
buildMatchMatrix[viralAccessionBatch_] := Map[
Function[{primerName},
Function[{viralAccession},
Boole[StringContainsQ[
accessionToViralSequence[viralAccession], primerToForwardPattern[primerName]
]]
] /@ viralAccessionBatch
],
Keys[primerToForwardPattern]
];
Column[Map[
ArrayPlot[buildMatchMatrix[#], Mesh -> True, Frame -> True, FrameTicks -> (listToTickLabels /@ {Keys[primerToForwardPattern], Rotate[#, 90 Degree] & /@ #}), ImageSize -> {Automatic, 220}] &,
Partition[Keys[accessionToViralSequence], UpTo[40]]
]]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/5bd172f1959ca135.png)

![originalReferenceSequence = Normal[ResourceData[
"Genetic Sequences for the SARS-CoV-2 Coronavirus"][
Select[#Accession === "NC_045512" &]]][[1]]["Sequence"];
sarsCoV2PrimerNames = Normal[ResourceData["CDC Primers for SARS-CoV-2 Research"][
Select[StringStartsQ[#Name, "2019"] &]][ReverseSortBy["Name"]][
All, "Name"]];
NumberLinePlot[
Interval @@@ StringPosition[originalReferenceSequence, primerToForwardPattern[#]] & /@ sarsCoV2PrimerNames,
PlotLegends -> sarsCoV2PrimerNames, PlotRange -> {28000, 29500}, AxesLabel -> "base pairs", PlotLabel -> "Primer Position on Reference SARS-CoV-2 Sequence"
]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/780451b8be1b419c.png)
![primerToForwardPattern = <|
Rule @@@ Normal[Values /@ ResourceData["CDC Primers for SARS-CoV-2 Research"][[
All, {"Name", "Forward Matching Pattern"}]]]|>;
findPosition[primerName_] := Interval[StringPosition[originalReferenceSequence, primerToForwardPattern[primerName]][[1]]];
Dataset[Module[{forwardPrimer = "2019-nCoV_N" <> # <> "-F", reversePrimer = "2019-nCoV_N" <> # <> "-R"},
<|"Forward Primer" -> forwardPrimer, "Reverse Primer" -> reversePrimer, "Amplicon Size" -> Min[findPosition[reversePrimer] - findPosition[forwardPrimer]]|>
] & /@ (ToString /@ Range[3])]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/5f2dac1637624024.png)
![ResourceData["CDC Primers for SARS-CoV-2 Research"][
All, {"Name", "Sequence String"}][All, Append[#, <|
"Genome Matches" -> GenomeLookup[
StringReplace[#["Sequence String"], "Y" -> "C"]]|>] &]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/3a0e815c987acccd.png)
![cgPercent[seqStr_] := With[{tally = Tally[Characters[seqStr]], totFunc = Total[Last /@ #] &},
Quantity[
100.*totFunc[Select[tally, MemberQ[{"C", "G"}, First[#]] &]]/
totFunc[tally], "Percent"]];
ResourceData["CDC Primers for SARS-CoV-2 Research"][
All, {"Name", "Sequence String"}][All, Append[#, <|"CG Percentage" -> cgPercent[#["Sequence String"]], "CG at 3'?" -> StringEndsQ[#["Sequence String"], "C" | "G"]|>] &][
SortBy["CG Percentage"]]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/221e6de6b9932775.png)
![tempApprox[seqStr_] := With[{countAssoc = <|Rule @@@ Tally[Characters[seqStr]]|>},
(2 Total[countAssoc /@ {"A", "T"}]) + (4 Total[
countAssoc /@ {"C", "G"}])
];
ResourceData["CDC Primers for SARS-CoV-2 Research"][
All, {"Name", "Sequence String"}][All, Append[#, <|
"Annealing Temp." -> tempApprox[#["Sequence String"]]|>] &][
SortBy["Name"]]](https://www.wolframcloud.com/obj/resourcesystem/images/f9f/f9fe12c8-797b-473f-9263-3008e5a3d3e4/5a13483cc2db1984.png)