Wolfram Data Repository
Immediate Computable Access to Curated Contributed Data
Medical records of patients infected with novel coronavirus COVID-19 (This data was imported and made computable on August 31, 2020.)
Patient record including age, sex, location, date of onset, symptoms, travel history, chronic diseases, and date of discharge or death.
The primary content includes medical records with patient characteristics information, but excluding the records in which only location information is available.
Records that contain only location information are accessible in the sub-element set ResourceData["Patient Medical Data for Novel Coronavirus COVID-19", "ReportedRecordCountByLocation"].
Retrieve the resource:
| In[1]:= |
| Out[1]= |  |
Retrieve the default content, which includes medical records with patient characteristics information (excluding the records in which only location information is available):
| In[2]:= |
| Out[2]= | 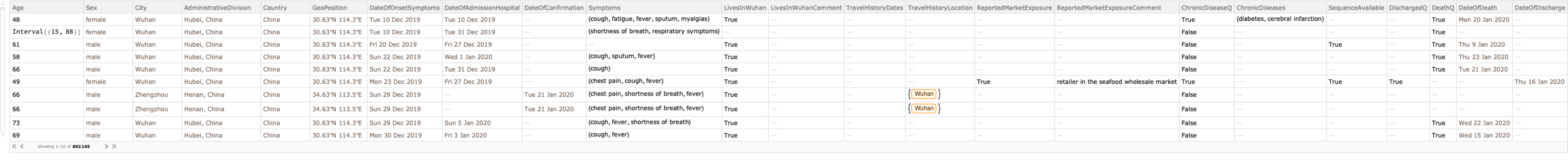 |
Retrieve counts of additional cases where only location and confirmation date is available:
| In[3]:= |
| Out[3]= | 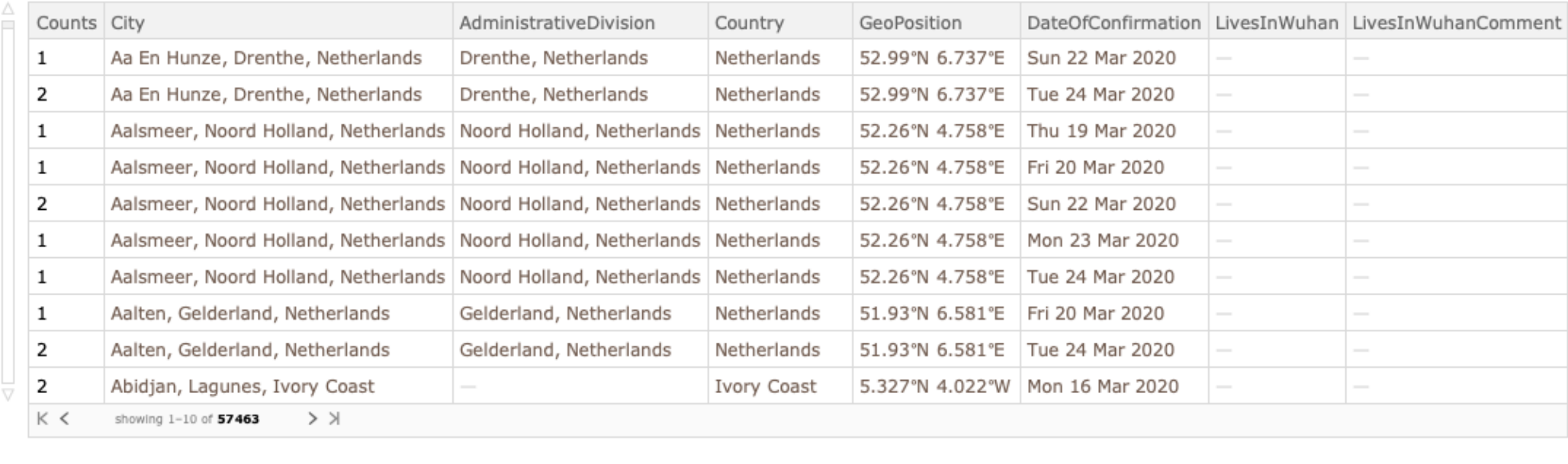 |
Combine these two datasets into one:
| In[4]:= | ![alldataset = Dataset[KeyUnion[
Join[Normal[
ResourceData[
"Patient Medical Data for Novel Coronavirus COVID-19"]], Flatten@Normal[
locationcountDataset[All, Table[Rest[#], #Counts] &]]]]];](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/07614002f84fab5f.png) |
Latest update date:
| In[5]:= |
| Out[5]= |
Compare the age distributions between male and female patients:
| In[6]:= | ![PairedHistogram[Sequence @@ (Most@Normal@#[Values]), ChartLabels -> Most@Normal@#[Keys], Sequence[
PlotLabel -> "age distribution", PlotTheme -> "Business"]] &@
ResourceData["Patient Medical Data for Novel Coronavirus COVID-19"][
GroupBy["Sex"], Select[NumberQ[#Age] &], "Age"]](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/45a0504d07c21633.png) |
| Out[6]= | 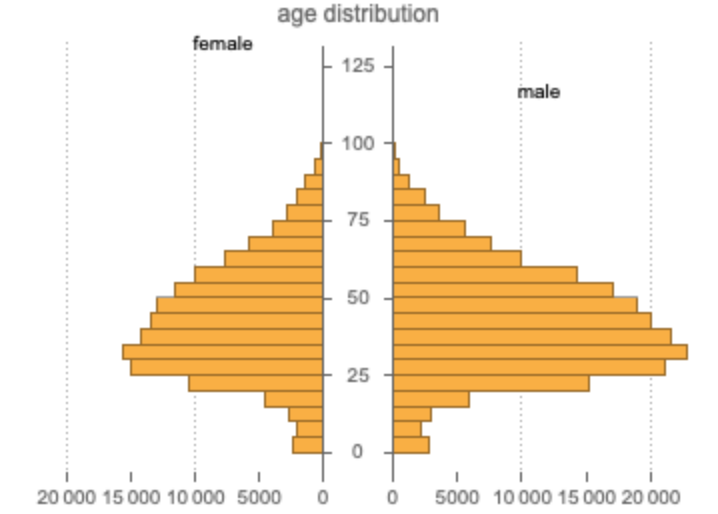 |
Plot the disease caused events including the onset of symptoms and the dates of confirmation or discharge for individual patients:
| In[7]:= | ![timelines = ResourceData["Patient Medical Data for Novel Coronavirus COVID-19"][
Select[(! MissingQ[#DateOfDeath] || ! MissingQ[#DateOfDischarge]) && ! MissingQ[#DateOfOnsetSymptoms] && ! MissingQ[#DateOfConfirmation] && ! MissingQ[#Age] &], {"DateOfOnsetSymptoms", "DateOfAdmissionHospital", "DateOfConfirmation", "DateOfDischarge", "DateOfDeath", "Age", "Sex"}];](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/42b5f93d4379fb49.png) |
| In[8]:= | ![Grid[Partition[
Labeled[TimelinePlot[Take[#, 5], Sequence[
PlotRange -> {"Dec 20 2019", "Feb 21 2020"}, ImageSize -> 220]],
StringTemplate["`Sex`, `Age` yrs old"][#], Bottom, LabelStyle -> Directive[FontFamily -> "Helvetica"]] & /@ Normal[Sort[timelines]][[;; 6]], 3], Alignment -> {{Left, Left}, {Bottom, Bottom}}]](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/4a7120f7c5560143.png) |
| Out[8]= | 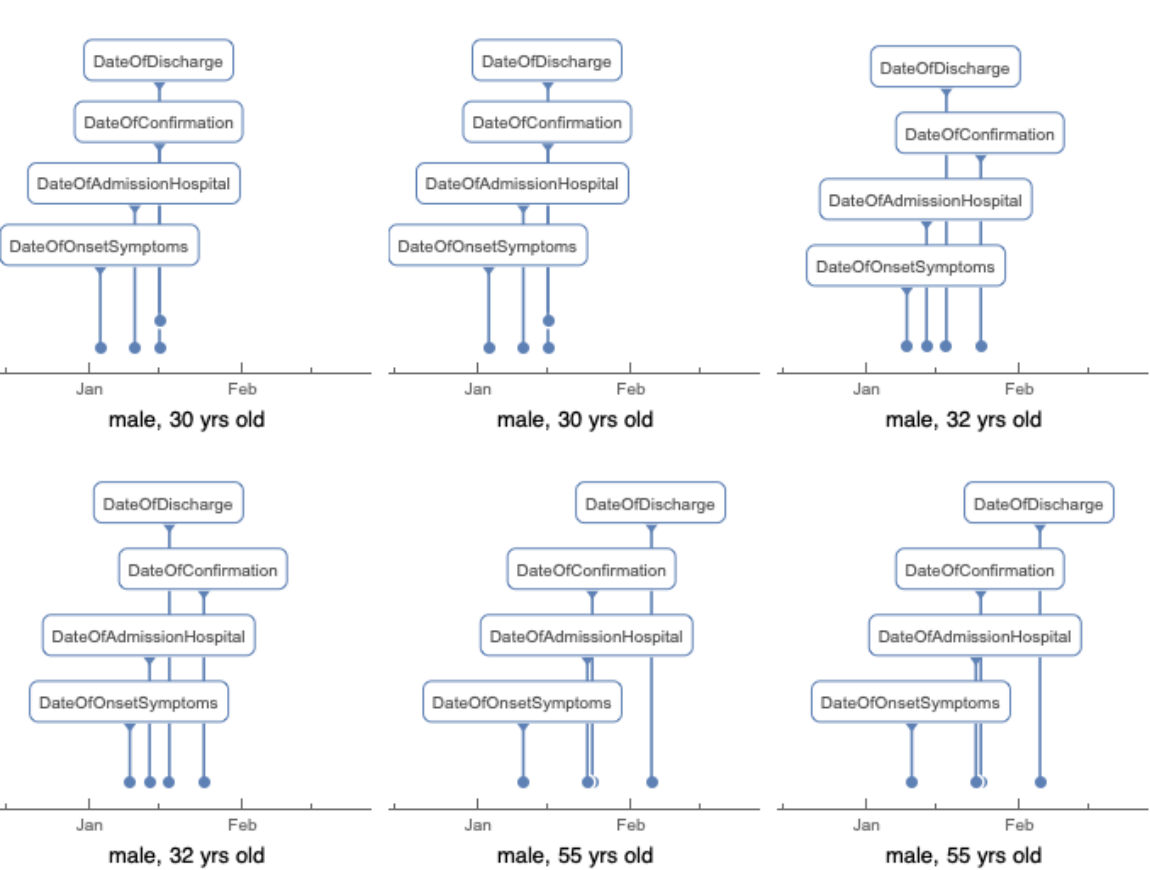 |
Explore links between the incidence of chronic diseases and deaths:
| In[9]:= | ![diseases = ResourceData["Patient Medical Data for Novel Coronavirus COVID-19"][
Select[! MissingQ[#ChronicDiseaseQ] && ! MissingQ[#DeathQ] &], {"ChronicDiseaseQ", "DeathQ", "ChronicDiseases"}]](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/4bad644da2c5babb.png) |
| Out[9]= | 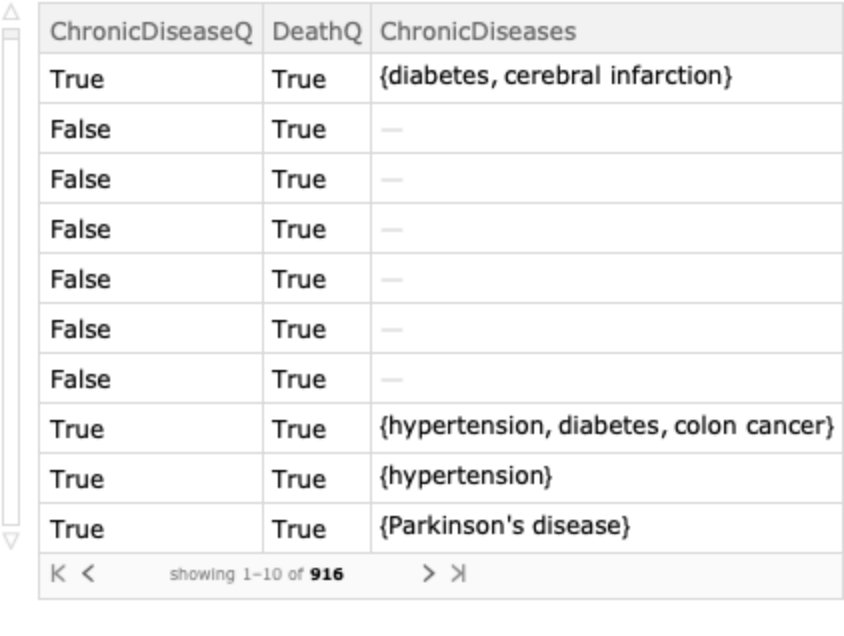 |
Find the top chronic conditions:
| In[10]:= |
| Out[10]= | 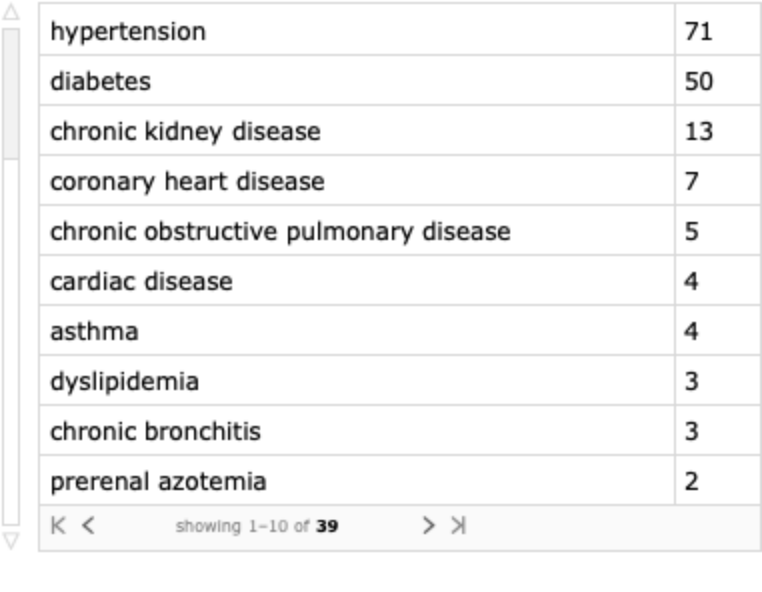 |
Find locations that infected individuals recently visited:
| In[11]:= | ![travelPath = GeoGraphics[{Red, Thickness[.001], Arrowheads[0.015], ResourceData[
"Patient Medical Data for Novel Coronavirus COVID-19"][
Select[(FreeQ[#TravelHistoryLocation, _Missing] && ! MissingQ[#City]) &], Arrow@GeoPath@Append[#TravelHistoryLocation, #City] &]}, GeoRange -> "World"]](https://www.wolframcloud.com/obj/resourcesystem/images/732/732a817d-f81e-4eca-a562-3323ba36145a/410c1a195bd97464.png) |
| Out[11]= |  |
Wolfram Research, "Patient Medical Data for Novel Coronavirus COVID-19" from the Wolfram Data Repository (2020) https://doi.org/10.24097/wolfram.11224.data